6PM9
 
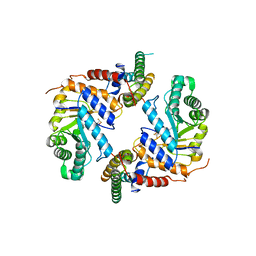 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
7WC3
 
 | | X-ray structure of the human adipocyte fatty acid-binding protein complexed with the fluorescent probe HA728 | | Descriptor: | (3S)-1-(4-nitro-2,1,3-benzoxadiazol-7-yl)piperidine-3-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M, Sugiyama, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of the human adipocyte fatty acid-binding protein complexed with the fluorescent probe HA728
To Be Published
|
|
6V51
 
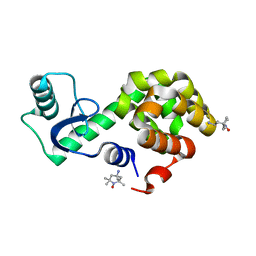 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
5AA2
 
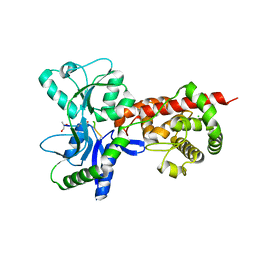 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. | | Descriptor: | CHLORIDE ION, MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, N-ACETYLGLUCOSAMINE-1,6-ANHYDRO-N-ACETYLMURAMIC ACID L-ALA-D-GLU-M-DAP-D-ALA-D-ALA | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
6AUX
 
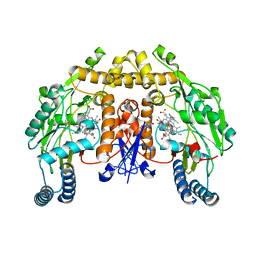 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(2-(5-Fluoro-3'-((methylamino)methyl)-[1,1'-biphenyl]-3-yl)ethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-fluoro-3'-[(methylamino)methyl][1,1'-biphenyl]-3-yl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6PN4
 
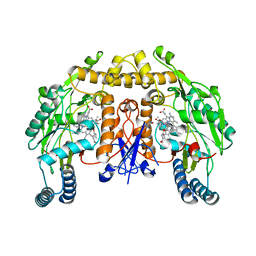 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclopropylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclopropylmethoxy)phenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8Q1H
 
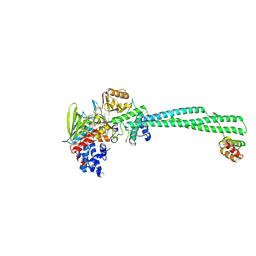 | | LSD1 Y391K-CoREST bound to Histone H3 N-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
6AV7
 
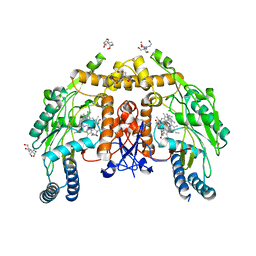 | | Structure of human endothelial nitric oxide synthase heme domain in complex with HW69 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
8BJH
 
 | | chimera of the inactive ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, with the double mutation K3528M and K3535I, fused to a proline-Rich-Domain (PRD) and profilin, bound to Latrunculin B-ADP-Mg-actin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
6PNF
 
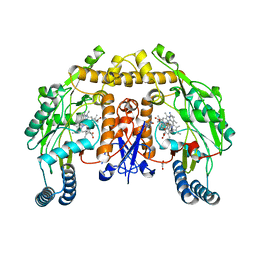 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-ethoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-ethoxyphenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5AA1
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide | | Descriptor: | CHLORIDE ION, MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, N-ACETYLGLUCOSAMINE-1,6-ANHYDRO-N-ACETYLMURAMIC ACID L-ALA-D-GLU-M-DAP-D-ALA-D-ALA | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
6BGB
 
 | |
8BBQ
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, GLYCEROL, ... | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
7WD6
 
 | | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with hexanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXANOIC ACID, ... | | Authors: | Sugiyama, S, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with hexanoic acid
To Be Published
|
|
6V5P
 
 | | EGFR(T790M/V948R) in complex with LN2725 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-(3-methoxypropyl)-1H-imidazol-5-yl]-2-phenyl-3H-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
5AL6
 
 | |
6POC
 
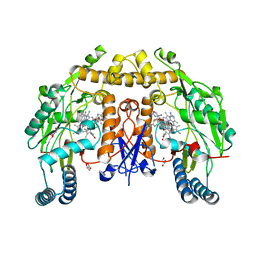 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(oxazol-4-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 7-{3-(aminomethyl)-4-[(1,3-oxazol-4-yl)methoxy]phenyl}-4-methylquinolin-2-amine, GLYCEROL, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7OPN
 
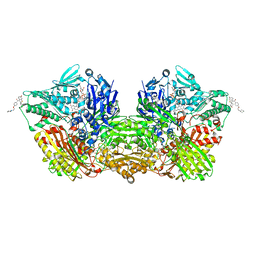 | | Human Aldehyde Oxidase SNP R1231H in complex with Raloxifene | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Aldehyde oxidase, DIMETHYL SULFOXIDE, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
7W3N
 
 | | Crystal structure of Ufm1 fused to UFBP1 UFIM | | Descriptor: | UFBP1 peptide,Ubiquitin-fold modifier 1 | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
6MJ8
 
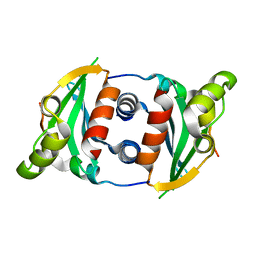 | |
8B39
 
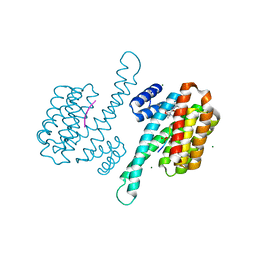 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080299) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[(2~{S},6~{R})-4-[(4-chlorophenyl)amino]-2,6-dimethyl-oxan-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8Q1G
 
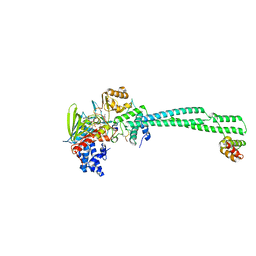 | | LSD1-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
6POW
 
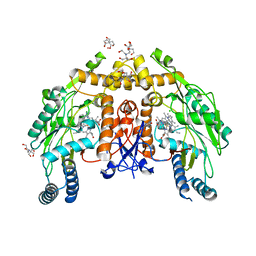 | | Structure of human endotheial nitric oxide synthase heme domain in complex with 7-(5-(Aminomethyl)pyridin-3-yl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[5-(aminomethyl)pyridin-3-yl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6AXN
 
 | | F95C Epi-isozizaene synthase | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Blank, P.N, Barrow, G.H, Christianson, D.W. | | Deposit date: | 2017-09-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substitution of Aromatic Residues with Polar Residues in the Active Site Pocket of epi-Isozizaene Synthase Leads to the Generation of New Cyclic Sesquiterpenes.
Biochemistry, 56, 2017
|
|
8BO1
 
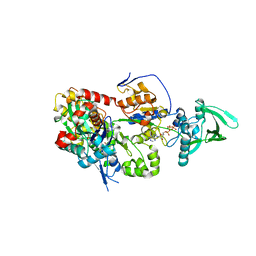 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
