6GTC
 
 | | Transition state structure of Cpf1(Cas12a) I1 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Mesa, P, Montoya, G. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTE
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I3 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), ... | | Authors: | Montoya, G, Mesa, P, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
4PQL
 
 | | N-Terminal domain of DNA binding protein | | Descriptor: | 1,2-ETHANEDIOL, Truncated replication protein RepA | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N.K. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6GTG
 
 | | Transition state structure of Cpf1(Cas12a) I4 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (32-MER), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), ... | | Authors: | Mesa, P, Montoya, G, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
3DLH
 
 | | Crystal structure of the guide-strand-containing Argonaute protein silencing complex | | Descriptor: | ACETIC ACID, Argonaute, DNA (5'-D(DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DAP*DTP*DAP*DGP*DT)-3'), ... | | Authors: | Wang, Y, Sheng, G, Patel, D.J. | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the guide-strand-containing argonaute silencing complex.
Nature, 456, 2008
|
|
3AZE
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K64Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZG
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K115Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
6GTF
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I5 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*A)-3'), ... | | Authors: | Montoya, G, Mesa, P, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTD
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I2 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Montoya, G, Mesa, P. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
5VHE
 
 | | DHX36 in complex with the c-Myc G-quadruplex | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36, DNA (5'-D(*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*TP*TP*TP*TP*T)-3'), POTASSIUM ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.793 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
3E7L
 
 | | Crystal structure of sigma54 activator NtrC4's DNA binding domain | | Descriptor: | Transcriptional regulator (NtrC family), ZINC ION | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
8HB2
 
 | |
8HBB
 
 | |
7MXW
 
 | |
7MXX
 
 | |
8GXV
 
 | |
7LYC
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A Lys13 and Lys15 in complex with BARD1 (residues 415-777) | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-06-16 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
7KLP
 
 | | Crystal structure of a three-tetrad, parallel, K+ stabilized human telomeric G-quadruplex | | Descriptor: | HEXAETHYLENE GLYCOL, POTASSIUM ION, telomeric DNA | | Authors: | Yatsunyk, L.A, Li, K.S, Chen, E.V. | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Water spines and networks in G-quadruplex structures.
Nucleic Acids Res., 49, 2021
|
|
7MI9
 
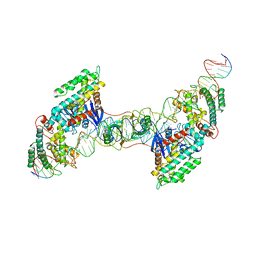 | | Full integration complex of Cas1/Cas2 from Cas4-containing system | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (5'-D(P*CP*GP*GP*AP*AP*AP*AP*GP*AP*GP*CP*C)-3'), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7MIB
 
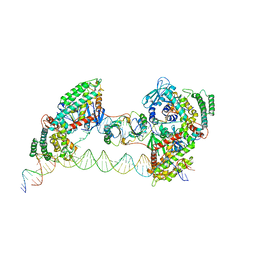 | |
1E7L
 
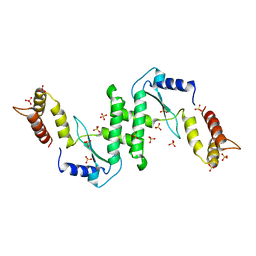 | | Endonuclease VII (EndoVII) N62D mutant from phage T4 | | Descriptor: | RECOMBINATION ENDONUCLEASE VII, SULFATE ION, ZINC ION | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-29 | | Release date: | 2001-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
5XF5
 
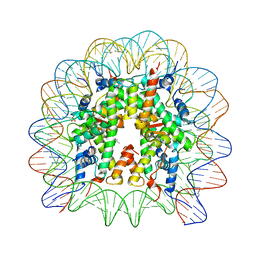 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,S-configuration) | | Descriptor: | (1S,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
4MKW
 
 | |
