6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6UAB
 
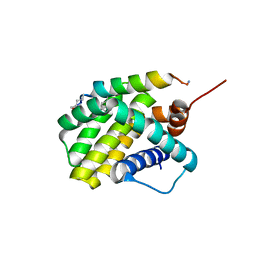 | |
8E58
 
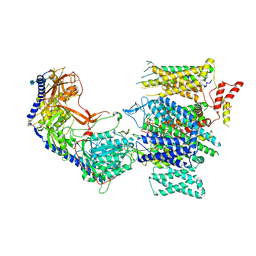 | | Rabbit L-type voltage-gated calcium channel Cav1.1 in the presence of Amiodarone and 1 mM MNI-1 at 3.0 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
6QHV
 
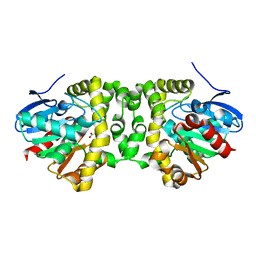 | | Time resolved structural analysis of the full turnover of an enzyme - 100 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
4BD8
 
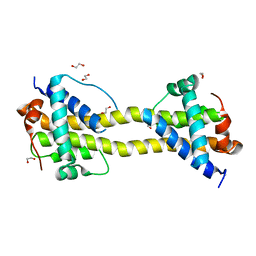 | | Bax domain swapped dimer induced by BimBH3 with CHAPS | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BAX, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4AV6
 
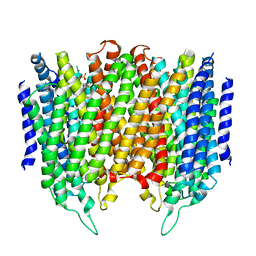 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase at 4 A in complex with phosphate and magnesium | | Descriptor: | K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kajander, T, Kellosalo, J, Kogan, K, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
6QKS
 
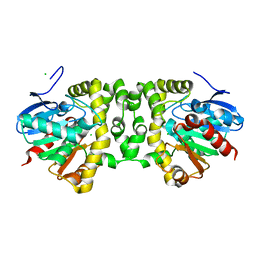 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6UCU
 
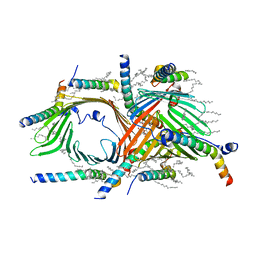 | | Cryo-EM structure of the mitochondrial TOM complex from yeast (dimer) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Park, E, Tucker, K. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TXS
 
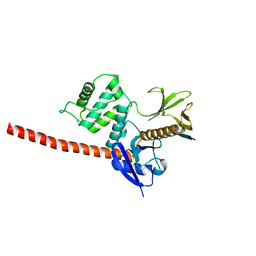 | | The structure of the FERM domain and helical linker of human moesin bound to a CD44 peptide | | Descriptor: | CD44 antigen, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
4AW6
 
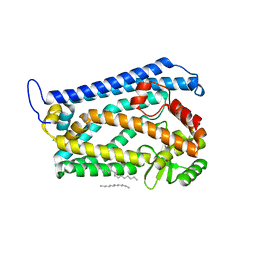 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 (FACE1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX PRENYL PROTEASE 1 HOMOLOG, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
6QLT
 
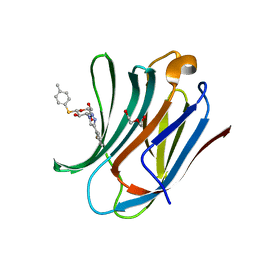 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-7 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, DI(HYDROXYETHYL)ETHER, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6U06
 
 | |
2KYK
 
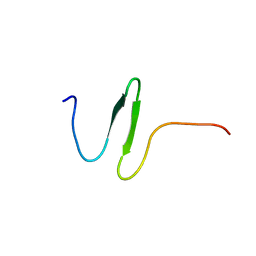 | | The sandwich region between two LMP2A PY motif regulates the interaction between AIP4WW2domain and PY motif | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Seo, M, Park, S, Seok, S, Kim, J, Cha, M, Lee, B. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The sandwich region between two LMP2A PY motif regulates the interaction between AIP4WW2domain and PY motif
To be Published
|
|
8EFH
 
 | | Helical reconstruction of the human cardiac actin-tropomyosin-myosin complex in complex with ADP-Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Doran, M.H, Lehman, W, Rynkiewicz, M.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes linked to ADP release from human cardiac myosin bound to actin-tropomyosin.
J.Gen.Physiol., 155, 2023
|
|
6QPB
 
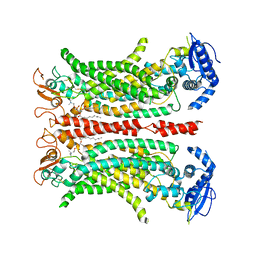 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6U37
 
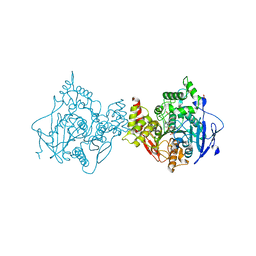 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
4BD7
 
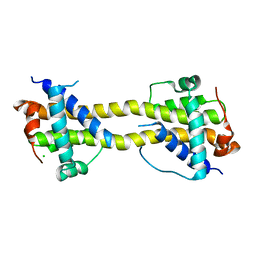 | | Bax domain swapped dimer induced by octylmaltoside | | Descriptor: | APOPTOSIS REGULATOR BAX, CHLORIDE ION, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
8DWI
 
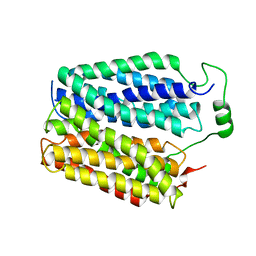 | |
4B0T
 
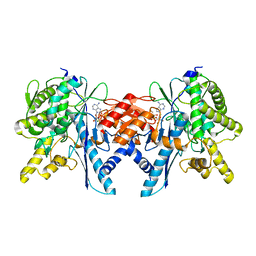 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
6U7E
 
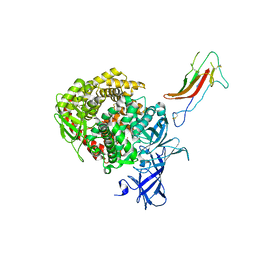 | | HCoV-229E RBD Class III in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6UC0
 
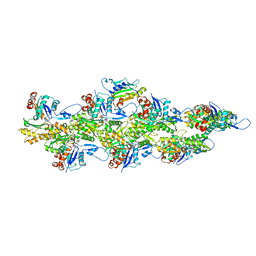 | | Isolated S3D-cofilin bound to an actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QSN
 
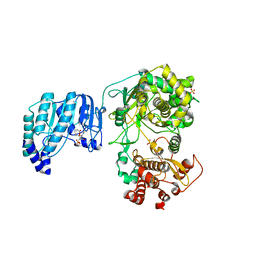 | | Crystal structure of Yellow fever virus polymerase NS5A | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Boura, E, Dubankova, A. | | Deposit date: | 2019-02-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses.
Antiviral Res., 169, 2019
|
|
4BP2
 
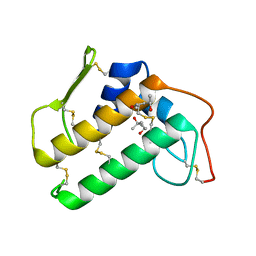 | | CRYSTALLOGRAPHIC REFINEMENT OF BOVINE PRO-PHOSPHOLIPASE A2 AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Finzel, B.C, Weber, P.C, Ohlendorf, D.H, Salemme, F.R. | | Deposit date: | 1990-09-07 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic refinement of bovine pro-phospholipase A2 at 1.6 A resolution.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
6U9H
 
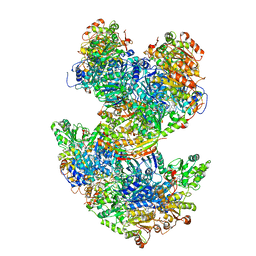 | | Arabidopsis thaliana acetohydroxyacid synthase complex | | Descriptor: | Acetolactate synthase small subunit 2, chloroplastic, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Low, Y.S, Rao, Z. | | Deposit date: | 2019-09-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of fungal and plant acetohydroxyacid synthases.
Nature, 586, 2020
|
|
4BXC
 
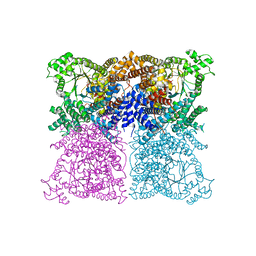 | | Resolving the activation site of positive regulators in plant phosphoenolpyruvate carboxylase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Schlieper, D, Foerster, K, Paulus, J.K, Groth, G. | | Deposit date: | 2013-07-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Resolving the Activation Site of Positive Regulators in Plant Phosphoenolpyruvate Carboxylase.
Mol.Plant, 7, 2014
|
|
