7CM9
 
 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
7CHS
 
 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CQD
 
 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA14 tag inserted between the residues 235 and 236 | | Descriptor: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | Authors: | Tamura-Sakaguchi, R, Aruga, R, Nogi, T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2MJ7
 
 | | Solution NMR structure of beta-adaptin appendage domain of human adaptor protein complex 4 subunit beta, Northeast Structural Genomics Consortium (NESG) Target HR8998C | | Descriptor: | AP-4 complex subunit beta-1 | | Authors: | Eletsky, A, Rotshteyn, D.J, Pederson, K, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-26 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of beta-adaptin appendage domain of human adaptor protein complex 4 subunit beta, Northeast Structural Genomics Consortium (NESG) Target HR8998C
To be Published
|
|
1NHU
 
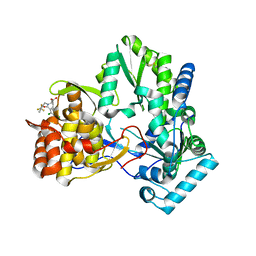 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
7CIN
 
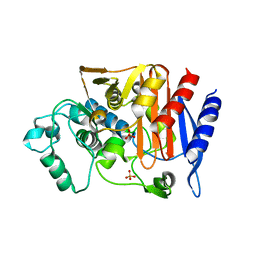 | |
1IVL
 
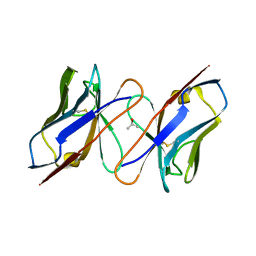 | |
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
8T5A
 
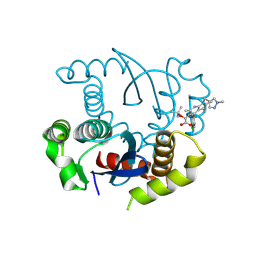 | |
2MC6
 
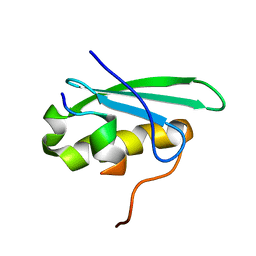 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement | | Descriptor: | DNA-directed RNA polymerase subunit beta', RNA polymerase inhibitor p7 | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-15 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
8TSZ
 
 | |
7CQC
 
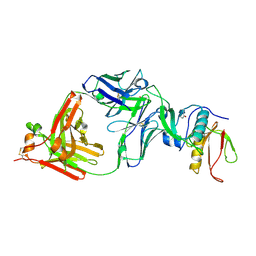 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA14 tag inserted between the residues 181 and 184 | | Descriptor: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | Authors: | Aruga, R, Tamura-Sakaguchi, R, Nogi, T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSY
 
 | |
2MF1
 
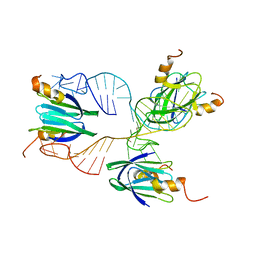 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MIZ
 
 | |
7CRA
 
 | |
6ALE
 
 | | A V-to-F substitution in SK2 channels causes Ca2+ hypersensitivity and improves locomotion in a C. elegans ALS model | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin-2, ... | | Authors: | Nam, Y.W, Zhang, M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A V-to-F substitution in SK2 channels causes Ca2+hypersensitivity and improves locomotion in a C. elegans ALS model.
Sci Rep, 8, 2018
|
|
2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | Descriptor: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
2M9W
 
 | | Solution NMR Structure of Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B | | Descriptor: | Transcription factor GATA-4, ZINC ION | | Authors: | Xu, X, Eletsky, A, Lee, D, Kohn, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B
To be Published
|
|
7C8E
 
 | | Crystal Structure of 14-3-3 epsilon with 9J10 peptide | | Descriptor: | 14-3-3 protein epsilon, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9J10 | | Authors: | Mathivanan, S, Sudhakar, S, Bairy, S, Kamariah, N, Venkitaraman, A. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Target identification for small-molecule discovery in the FOXO3a tumor-suppressor pathway using a biodiverse peptide library.
Cell Chem Biol, 28, 2021
|
|
7CR9
 
 | |
7C8H
 
 | | Ambient temperature structure of Bifidobacterium longum phosphoketolase with thiamine diphosphate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, MALONIC ACID, ... | | Authors: | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1JAV
 
 | |
