3SQ8
 
 | |
3N5E
 
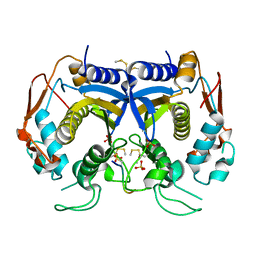 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | Descriptor: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P8R
 
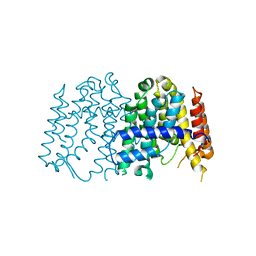 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE FROM Vibrio cholerae | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3SQ7
 
 | |
3OYR
 
 | | Crystal structure of polyprenyl synthase from Caulobacter crescentus CB15 complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, PYROPHOSPHATE 2-, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3P8L
 
 | | Crystal structure of polyprenyl synthase from Enterococcus faecalis V583 | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-14 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3P41
 
 | | Crystal structure of polyprenyl synthetase from pseudomonas fluorescens pf-5 complexed with magnesium and isoprenyl pyrophosphate | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-05 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3PDE
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Lactobacillus brevis atcc 367 complexed with isoprenyl diphosphate and magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Farnesyl-diphosphate synthase, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3EA1
 
 | | Crystal Structure of the Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
3Q2Q
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SUX
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI AND 3-(2-benzothiazolylthio)-1-propanesulfonic acid | | Descriptor: | 3-(2-BENZOTHIAZOLYLTHIO)-1-PROPANESULFONIC ACID, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Tellez-Valencia, A, Olivares-Illana, V, Hernandez-Santoyo, A, Perez-Montfort, R, Costas, M, Rodriguez-Romero, A, Tuena De Gomez-Puyou, M, Gomez-Puyou, A. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface.
J.Mol.Biol., 341, 2004
|
|
3Q4D
 
 | | Crystal structure of dipeptide epimerase from Cytophaga hutchinsonii complexed with Mg and dipeptide D-Ala-L-Ala | | Descriptor: | ALANINE, D-ALANINE, MAGNESIUM ION, ... | | Authors: | Lukk, T, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4U0A
 
 | | Hexameric HIV-1 CA in complex with CPSF6 peptide, P6 crystal form | | Descriptor: | CHLORIDE ION, Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0E
 
 | | Hexameric HIV-1 CA in complex with PF3450074 | | Descriptor: | CHLORIDE ION, Capsid protein p24, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0F
 
 | | Hexameric HIV-1 CA in Complex with BI-2 | | Descriptor: | (4S)-4-(4-hydroxyphenyl)-3-phenyl-4,5-dihydropyrrolo[3,4-c]pyrazol-6(1H)-one, 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
3E29
 
 | | X-Ray structure of the protein Q7WE92_BORBR from thioesterase superfamily. Northeast Structural Genomics Consortium Target BoR214A. | | Descriptor: | uncharacterized protein Q7WE92_BORBR | | Authors: | Kuzin, A.P, Chen, Y, Setharaman, J, Wang, D, Mao, L, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q7WE92_BORBR from thioesterase superfamily. Northeast Structural Genomics Consortium Target BoR214A.
To be Published
|
|
6DNK
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
4U0B
 
 | |
4U0D
 
 | | Hexameric HIV-1 CA in complex with Nup153 peptide, P212121 crystal form | | Descriptor: | CHLORIDE ION, Gag polyprotein, Nuclear pore complex protein Nup153 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
2LMZ
 
 | |
4ZMD
 
 | | C domain of staphylococcal protein A mutant - Q9W | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Deis, L.N, Oas, T.G. | | Deposit date: | 2015-05-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Suppression of conformational heterogeneity at a protein-protein interface.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5XOR
 
 | |
7QLB
 
 | | SMYD3 in complex with fragment FL06268 | | Descriptor: | 1-methylimidazole-4-sulfonamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2021-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening
Rsc Med Chem, 2024
|
|
7QNU
 
 | | SMYD3 in complex with fragment FL08619 | | Descriptor: | BENZOYL-FORMIC ACID, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2021-12-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening
Rsc Med Chem, 2024
|
|
7QNR
 
 | | SMYD3 in complex with fragment FL01791 | | Descriptor: | 3-propan-2-yl-1,2,4-thiadiazol-5-amine, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2021-12-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening
Rsc Med Chem, 2024
|
|
