6AUP
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | GLYCEROL, Potassium channel toxin gamma-KTx 2.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
9IXG
 
 | |
9IXE
 
 | |
3G16
 
 | |
3BCA
 
 | |
6MM4
 
 | |
3BCB
 
 | |
6AV8
 
 | |
6AU7
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | GLYCEROL, Potassium channel toxin gamma-KTx 2.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-08-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6AVA
 
 | |
3BC8
 
 | | Crystal structure of mouse selenocysteine synthase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, O-phosphoseryl-tRNA(Sec) selenium transferase | | Authors: | Ganichkin, O.M, Wahl, M.C. | | Deposit date: | 2007-11-12 | | Release date: | 2007-12-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of eukaryotic selenocysteine synthase.
J.Biol.Chem., 283, 2008
|
|
1UZW
 
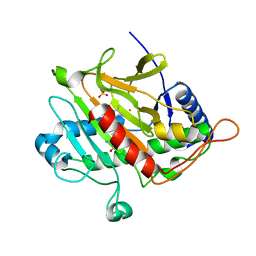 | | ISOPENICILLIN N SYNTHASE WITH L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-ISODEHYDROVALINE | | Descriptor: | D-(L-A-AMINOADIPOYL)-L-CYSTEINYL-D-ISODEHYDROVALINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Grummitt, A.R, Rutledge, P.J, Clifton, I.J, Baldwin, J.E. | | Deposit date: | 2004-03-17 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Active Site Mediated Elimination of Hydrogen Fluoride from a Fluorinated Substrate Analogue by Isopenicillin N Synthase
Biochem.J., 382, 2004
|
|
7VEO
 
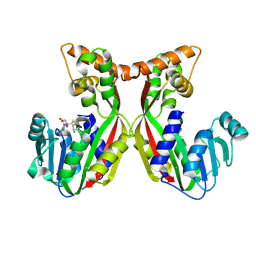 | | Crystal structure of juvenile hormone acid methyltransferase silkworm JHAMT isoform3 complex with S-Adenosyl-L-homocysteine | | Descriptor: | Methyltranfer_dom domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
6ATW
 
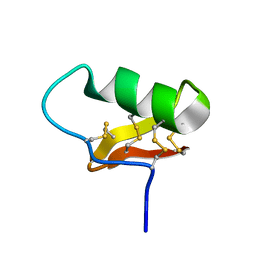 | |
4QEZ
 
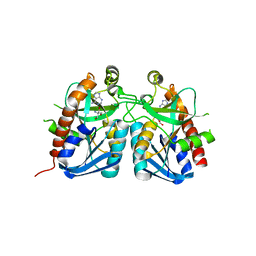 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Bacillus anthracis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE | | Authors: | Tarique, K.F, Devi, S, Abdul Rehman, S.A, Gourinath, S. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Bacillus anthracis
To be Published
|
|
6JLS
 
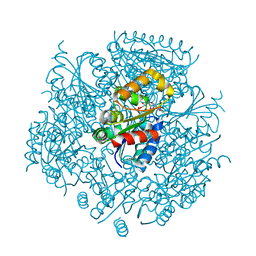 | | Crystal Structure of FMN-dependent Cysteine Decarboxylases TvaF from Thioviridamide Biosynthesis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative flavoprotein decarboxylase | | Authors: | Li, J, Lu, J, Wang, H, Zhu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the FMN-Dependent Cysteine Decarboxylase from Thioviridamide Biosynthesis.
Org.Lett., 21, 2019
|
|
3BBD
 
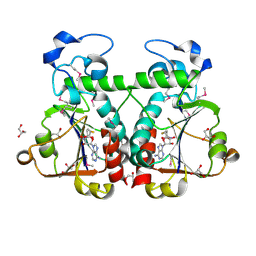 | | M. jannaschii Nep1 complexed with S-adenosyl-homocysteine | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
2BH2
 
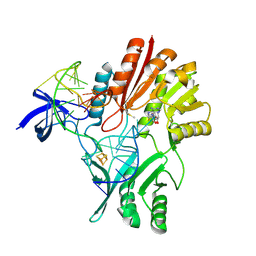 | | Crystal Structure of E. coli 5-methyluridine methyltransferase RumA in complex with ribosomal RNA substrate and S-adenosylhomocysteine. | | Descriptor: | 23S RIBOSOMAL RNA 1932-1968, 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2005-01-06 | | Release date: | 2005-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function
Cell(Cambridge,Mass.), 120, 2005
|
|
3FF2
 
 | |
9CEN
 
 | |
3FD0
 
 | |
4GMH
 
 | | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ACETATE ION | | Authors: | Brown, R.L, Anderson, B.F, Norris, G.E, Tyler, P.C, Evans, G.B, Sutherland-Smith, A.J. | | Deposit date: | 2012-08-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase
To be Published
|
|
1OH1
 
 | | Solution structure of staphostatin A form Staphylococcus aureus confirms the discovery of a novel class of cysteine proteinase inhibitors. | | Descriptor: | STAPHOSTATIN A | | Authors: | Dubin, G, Popowicz, G, Krajewski, M, Stec, J, Bochtler, M, Potempa, J, Dubin, A, Holak, T.A. | | Deposit date: | 2003-05-21 | | Release date: | 2003-11-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Biochemistry, 42, 2003
|
|
3W1H
 
 | |
3WCN
 
 | |
