6ZUV
 
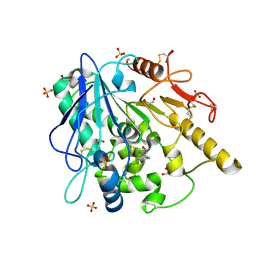 | | Notum fragment 286 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
6ZGV
 
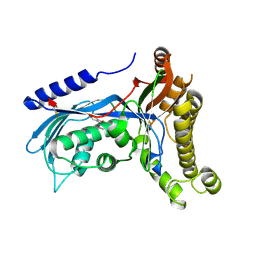 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 2-(4-chlorophenyl)-~{N}-pyrimidin-2-yl-ethanamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6ZFA
 
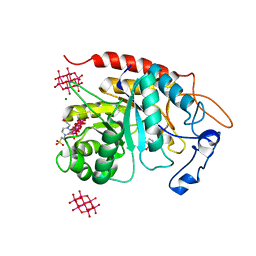 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with GlcIFG, alpha-1,2-mannobiose and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, 6-tungstotellurate(VI), ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YV4
 
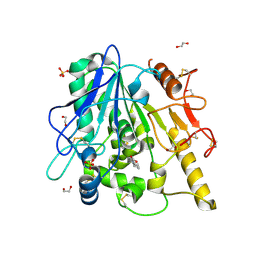 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 686 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | Authors: | Skerlova, J, Pachl, P, Rezacova, P. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
6PJK
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-29 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-isoleucyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
9RF2
 
 | | M. tuberculosis meets European Lead Factory: identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography: ELF5 | | Descriptor: | CHLORIDE ION, Monoacylglycerol lipase, [1-[6-[(4-methoxyphenyl)amino]pyrimidin-4-yl]pyrazol-4-yl]-[(3R)-3-oxidanylpyrrolidin-1-yl]methanone, ... | | Authors: | Riegler-Berket, L, Goedl, L, Oberer, M, Polidori, N. | | Deposit date: | 2025-06-04 | | Release date: | 2025-08-13 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | M. tuberculosis meets European lead factory:Identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography
Disease and Therapeutics, 1, 2025
|
|
6OPV
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPZ
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, I54L, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPT
 
 | | HIV-1 Protease NL4-3 V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
6OPX
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6ZGY
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | (2,5-dimethylphenyl) pyridine-4-carboxylate, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6PJL
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-95 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-alloisoleucyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Dybkov, O, Kastner, B, Luehrmann, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
7LPF
 
 | |
7LK0
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | Descriptor: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LTP
 
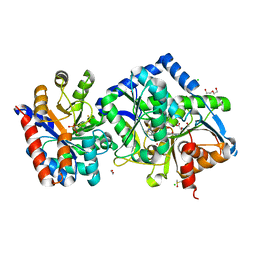 | | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and the product L-tryptophan at the enzyme beta-site at 1.47 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-02-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and the product L-tryptophan at the enzyme beta-site at 1.47 Angstrom resolution.
To be Published
|
|
7LV5
 
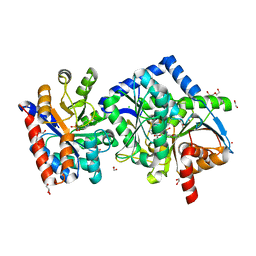 | | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and L-Histidine at the enzyme beta-site at 1.55 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-02-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and L-Histidine at the enzyme beta-site at 1.55 Angstrom resolution
To be Published
|
|
7LUT
 
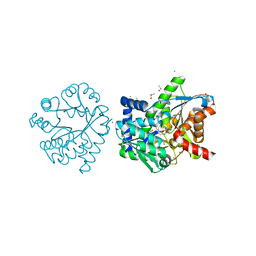 | |
5L0A
 
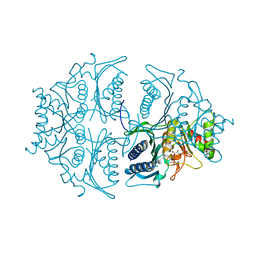 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
7LY8
 
 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with two molecules of N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the enzyme alpha-site, a single F6F molecule at the enzyme beta-site, and sodium ion at the metal coordination site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, CHLORIDE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-03-06 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with two molecules of N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the enzyme alpha-site, a single F6F molecule at the enzyme beta-site, and sodium ion at the metal coordination site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
9MTV
 
 | | Crystal structure of an apo-ADC-1 triple mutant | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, NITRATE ION | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-01-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of carbapenemase activity in the class C beta-lactamase ADC-1.
Mbio, 16, 2025
|
|
7LVX
 
 | |
6YKD
 
 | | Human Pim-1 kinase in complex with an inhibitor identified by virtual screening | | Descriptor: | ACETATE ION, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Schneider, P, Welin, M, Svensson, B, Walse, B, Schneider, G. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Virtual Screening and Design with Machine Intelligence Applied to Pim-1 Kinase Inhibitors.
Mol Inform, 39, 2020
|
|
