4ZVQ
 
 | |
5A95
 
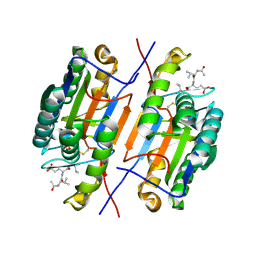
 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5A8N
 
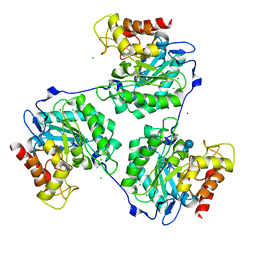
 | | Crystal structure of the native form of beta-glucanase SdGluc5_26A from Saccharophagus degradans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5A2K
 
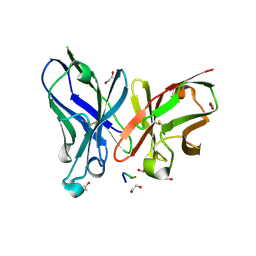 | | Crystal structure of scFv-SM3 in complex with APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5FML
 
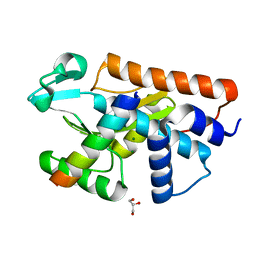 | | Crystal structure of the endonuclease from the PA subunit of influenza B virus bound to the PB2 subunit NLS peptide | | Descriptor: | GLYCEROL, MAGNESIUM ION, PA SUBUNIT OF INFLUENZA B POLYMERASE, ... | | Authors: | Guilligay, D, Gaudon, S, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
8GU3
 
 | |
4CE4
 
 | | 39S large subunit of the porcine mitochondrial ribosome | | Descriptor: | 16S Ribosomal RNA, ICT1, MRPL13, ... | | Authors: | Greber, B.J, Boehringer, D, Leitner, A, Bieri, P, Voigts-Hoffmann, F, Erzberger, J.P, Leibundgut, M, Aebersold, R, Ban, N. | | Deposit date: | 2013-11-08 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Architecture of the Large Subunit of the Mammalian Mitochondrial Ribosome.
Nature, 505, 2014
|
|
5YZ1
 
 | | Crystal structure of human Archease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein archease | | Authors: | Duan, S.Y, Li, J.X. | | Deposit date: | 2017-12-11 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of human archease, a key cofactor of tRNA splicing ligase complex.
Int.J.Biochem.Cell Biol., 122, 2020
|
|
3CC4
 
 | |
1T4N
 
 | | Solution structure of Rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
1T4O
 
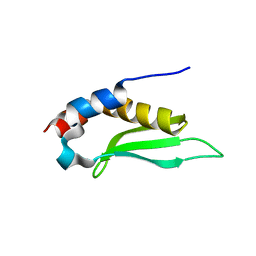 | | Crystal structure of rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares Jr, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
6P19
 
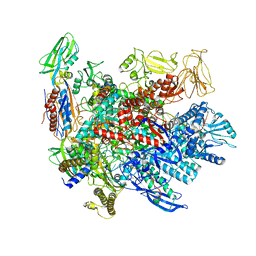 | | Q21 transcription antitermination complex: loaded complex | | Descriptor: | DNA (123-MER) fragment carrying phage-21 pR' promoter, pause element, and transcribed region, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P18
 
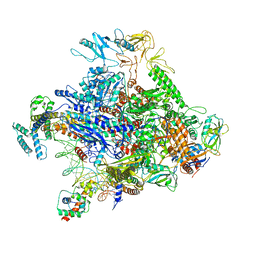 | | Q21 transcription antitermination complex: loading complex | | Descriptor: | DNA (67-MER) fragment carrying phage-21 pR' promoter and pause element, nontemplate strand, template strand, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XLL
 
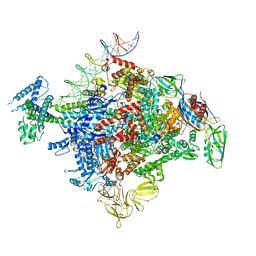 | | Cryo-EM structure of E. coli RNAP-promoter initial transcribing complex with 5-nt RNA transcript (RPitc-5nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
3RTX
 
 | |
7QUU
 
 | | Red1-Iss10 complex | | Descriptor: | NURS complex subunit red1, Uncharacterized protein C7D4.14c | | Authors: | Mackereth, C.D, Kadlec, J, Laroussi, H. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
3RER
 
 | | Crystal structure of E. coli Hfq in complex with AU6A RNA and ADP | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, W.W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperation of Escherichia coli Hfq hexamers in DsrA binding.
Genes Dev., 25, 2011
|
|
8IGS
 
 | | Cryo-EM structure of RNAP-promoter open complex at lambda promoter PRE | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
4D5Y
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 28S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em Structures of Ribosomal 80S Complexes with Termination Factors and Cricket Paralysis Virus Ires Reveal the Ires in the Translocated State
Mol.Cell, 57, 2015
|
|
8UO6
 
 | |
6D1R
 
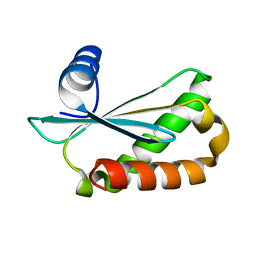 | | Structure of Staphylococcus aureus RNase P protein at 2.0 angstrom | | Descriptor: | Ribonuclease P protein component | | Authors: | Ha, L, Colquhoun, J, Noinaj, N, Das, C, Dunman, P, Flaherty, D.P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of the ribonuclease-P-protein subunit from Staphylococcus aureus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7MNX
 
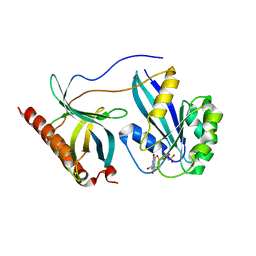 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 2 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO5
 
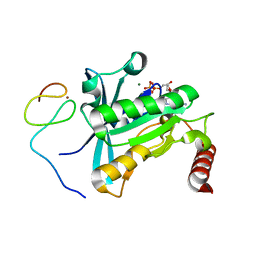 | | Crystal Structure of the ZnF4 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNS
 
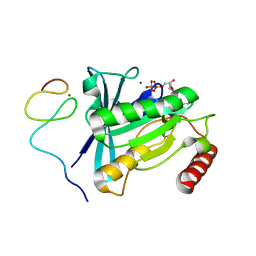 | | Crystal Structure of the ZnF4 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNV
 
 | | Crystal Structure of the ZnF8 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
