2J49
 
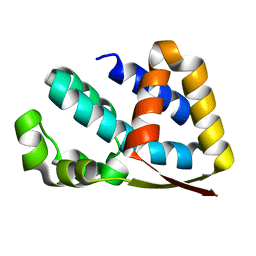 | | Crystal structure of yeast TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 5 | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
2J0V
 
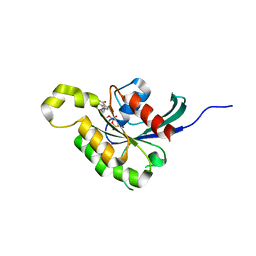 | | The crystal structure of Arabidopsis thaliana RAC7-ROP9: the first RAS superfamily GTPase from the plant kingdom | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAC-LIKE GTP-BINDING PROTEIN ARAC7 | | Authors: | Sormo, C.G, Leiros, I, Brembu, T, Winge, P, Os, V, Bones, A.M. | | Deposit date: | 2006-08-07 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Crystal Structure of Arabidopsis Thaliana Rac7/Rop9: The First Ras Superfamily Gtpase from the Plant Kingdom.
Phytochemistry, 67, 2006
|
|
2J4B
 
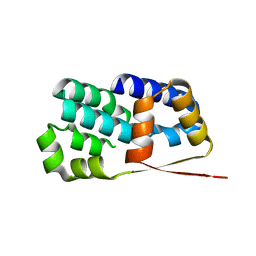 | | Crystal structure of Encephalitozoon cuniculi TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 72/90-100 KDA | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
1Z5R
 
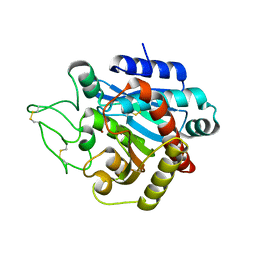 | | Crystal Structure of Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-03-18 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1ZG9
 
 | | Crystal Structure of 5-{[amino(imino)methyl]amino}-2-(sulfanylmethyl)pentanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 5-{[AMINO(IMINO)METHYL]AMINO}-2-(SULFANYLMETHYL)PENTANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
4C41
 
 | |
1H1J
 
 | |
4BGA
 
 | |
4BGB
 
 | |
4CX8
 
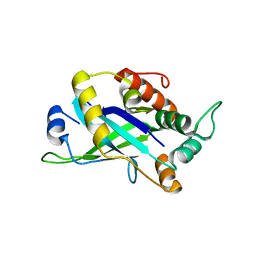 | | Monomeric pseudorabies virus protease pUL26N at 2.5 A resolution | | Descriptor: | PSEUDORABIES VIRUS PROTEASE | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease.
Plos Pathog., 11, 2015
|
|
2RTU
 
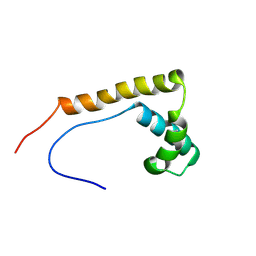 | | Solution structure of oxidized human HMGB1 A box | | Descriptor: | High mobility group protein B1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA.
Biochem.Biophys.Res.Commun., 441, 2013
|
|
2RUH
 
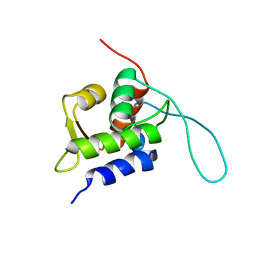 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
2VDY
 
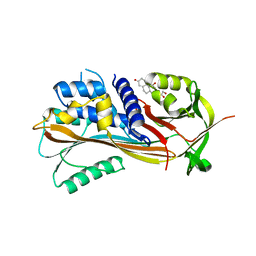 | | Crystal structure of the reactive loop cleaved Corticosteroid Binding Globulin complexed with Cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, CORTICOSTEROID-BINDING GLOBULIN | | Authors: | Zhou, A, Wei, Z, Read, R.J. | | Deposit date: | 2007-10-13 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The S-to-R Transition of Corticosteroid-Binding Globulin and the Mechanism of Hormone Release.
J.Mol.Biol., 380, 2008
|
|
2UZQ
 
 | | Protein Phosphatase, New Crystal Form | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U. | | Deposit date: | 2007-05-01 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
To be Published
|
|
2VDX
 
 | |
2VBQ
 
 | | Structure of AAC(6')-Iy in complex with bisubstrate analog CoA-S- monomethyl-acetylneamine. | | Descriptor: | (3R,9Z)-17-[(2R,3S,4R,5R,6R)-5-amino-6-{[(1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl]oxy}-3,4-dihydroxytetrahydro-2H-pyran-2-yl]-3-hydroxy-2,2-dimethyl-4,8,15-trioxo-12-thia-5,9,16-triazaheptadec-9-en-1-yl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, AMINOGLYCOSIDE 6'-N-ACETYLTRANSFERASE, GLYCEROL, ... | | Authors: | Vetting, M.W, Magalhaes, M.L, Freiburger, L, Gao, F, Auclair, K, Blanchard, J.S. | | Deposit date: | 2007-09-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and Structural Analysis of Bisubstrate Inhibition of the Salmonella Enterica Aminoglycoside 6'-N-Acetyltransferase.
Biochemistry, 47, 2008
|
|
2VQI
 
 | | Structure of the P pilus usher (PapC) translocation pore | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE USHER PROTEIN PAPC | | Authors: | Remaut, H, Tang, C, Henderson, N.S, Pinkner, J.S, Wang, T, Hultgren, S.J, Thanassi, D.G, Li, H, Waksman, G. | | Deposit date: | 2008-03-16 | | Release date: | 2008-05-27 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Fiber formation across the bacterial outer membrane by the chaperone/usher pathway.
Cell, 133, 2008
|
|
2VRG
 
 | | Structure of human MCFD2 | | Descriptor: | CALCIUM ION, MULTIPLE COAGULATION FACTOR DEFICIENCY PROTEIN 2 | | Authors: | Guy, J.E, Wigren, E, Svard, M, Hard, T, Lindqvist, Y. | | Deposit date: | 2008-04-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | New insights into multiple coagulation factor deficiency from the solution structure of human MCFD2.
J. Mol. Biol., 381, 2008
|
|
2VVY
 
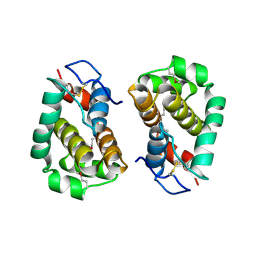 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2R7E
 
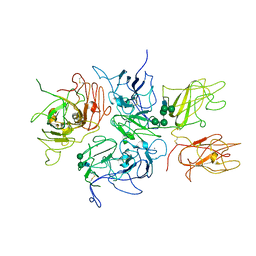 | | Crystal Structure Analysis of Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Stoddard, B.L, Shen, B.W. | | Deposit date: | 2007-09-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The tertiary structure and domain organization of coagulation factor VIII.
Blood, 111, 2008
|
|
2VY3
 
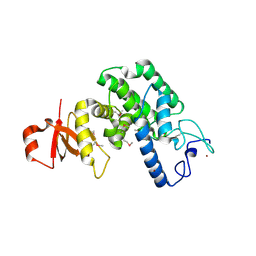 | | Type IV secretion system effector protein BepA | | Descriptor: | CELL FILAMENTATION PROTEIN, CHLORIDE ION, NICKEL (II) ION | | Authors: | Palanivelu, D.V, Schirmer, T. | | Deposit date: | 2008-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fic Domain Catalyzed Adenylylation: Insight Provided by the Structural Analysis of the Type Iv Secretion System Effector Bepa.
Protein Sci., 20, 2011
|
|
2R0L
 
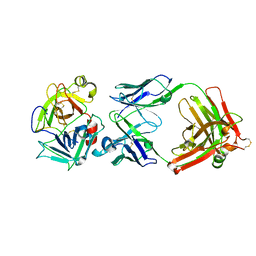 | | Short Form HGFA with Inhibitory Fab75 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0K
 
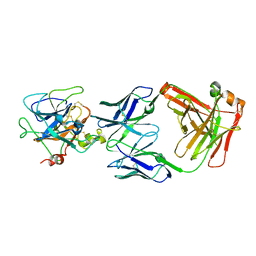 | | Protease domain of HGFA with inhibitor Fab58 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain of Fab58, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2VXH
 
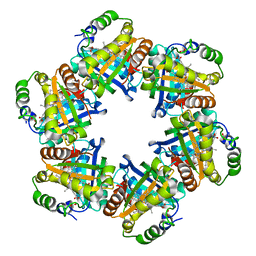 | | The crystal structure of chlorite dismutase: a detox enzyme producing molecular oxygen | | Descriptor: | CARBONATE ION, CHLORITE DISMUTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | De Geus, D.C, Thomassen, E.A.J, Hagedoorn, P.L, Pannu, N.S, Abrahams, J.P. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Chlorite Dismutase, a Detoxifying Enzyme Producing Molecular Oxygen
J.Mol.Biol., 387, 2009
|
|
2WCI
 
 | | Structure of E. coli monothiol glutaredoxin GRX4 homodimer | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTAREDOXIN-4, GLUTATHIONE, ... | | Authors: | Iwema, T, Picchiocci, A, Traore, D.A.K, Ferrer, J.-L, Chauvat, F, Jacquamet, L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Delivery of the Intact [Fe2S2] Cluster by Monothiol Glutaredoxin.
Biochemistry, 48, 2009
|
|
