5MDW
 
 | | Structure of ArfA(A18T) and RF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDV
 
 | | Structure of ArfA and RF2 bound to the 70S ribosome (accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDY
 
 | | Structure of ArfA and TtRF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDZ
 
 | | Structure of the 70S ribosome (empty A site) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
4C48
 
 | | Crystal structure of AcrB-AcrZ complex | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, DARPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Du, D, James, N, Klimont, E, Luisi, B.F. | | Deposit date: | 2013-09-02 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the AcrAB-TolC multidrug efflux pump.
Nature, 509, 2014
|
|
4CDI
 
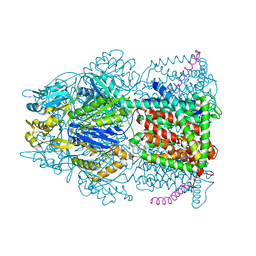 | | Crystal structure of AcrB-AcrZ complex | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, PREDICTED PROTEIN | | Authors: | Du, D, James, N, Klimont, E, Luisi, B.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the Acrab-Tolc Multidrug Efflux Pump.
Nature, 509, 2014
|
|
2J49
 
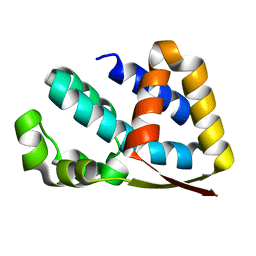 | | Crystal structure of yeast TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 5 | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
2J4B
 
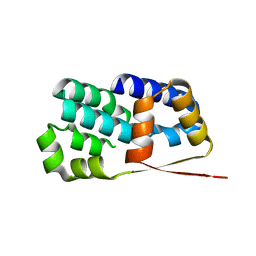 | | Crystal structure of Encephalitozoon cuniculi TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 72/90-100 KDA | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
