1OJG
 
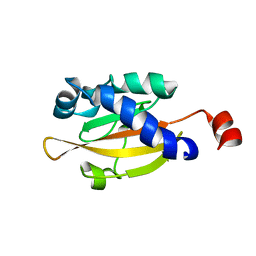 | | Sensory domain of the membraneous two-component fumarate sensor DcuS of E. coli | | Descriptor: | SENSOR PROTEIN DCUS | | Authors: | Pappalardo, L, Janausch, I.G, Vijayan, V, Zientz, E, Junker, J, Peti, W, Zweckstetter, M, Unden, G, Griesinger, C. | | Deposit date: | 2003-07-10 | | Release date: | 2003-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli.
J. Biol. Chem., 278, 2003
|
|
5H28
 
 | |
1ZM0
 
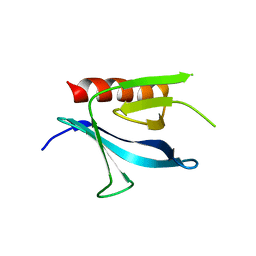 | | Crystal Structure of the Carboxyl Terminal PH Domain of Pleckstrin To 2.1 Angstroms | | Descriptor: | Pleckstrin | | Authors: | Jackson, S.G, Zhang, Y, Zhang, K, Summerfield, R, Haslam, R.J, Junop, M.S. | | Deposit date: | 2005-05-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the carboxy-terminal PH domain of pleckstrin at 2.1 Angstroms.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1YWR
 
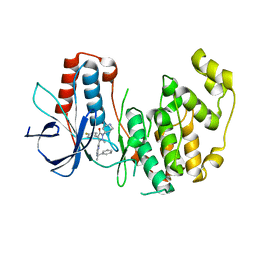 | | Crystal Structure Analysis of inactive P38 kinase domain in complex with a Monocyclic Pyrazolone Inhibitor | | Descriptor: | 4-(4-FLUOROPHENYL)-1-METHYL-5-(2-{[(1S)-1-PHENYLETHYL]AMINO}PYRIMIDIN-4-YL)-2-PIPERIDIN-4-YL-1,2-DIHYDRO-3H-PYRAZOL-3-ONE, Mitogen-activated protein kinase 14 | | Authors: | Golebiowski, A, Townes, J.A, Laufersweiler, M.J, Brugel, T.A, Clark, M.P, Clark, C.M, Djung, J.F, Laughlin, S.K, Sabat, M.P, Bookland, R.G, Vanrens, J.C, De, B, Hsieh, L.C, Janusz, M.J, Walter, R.L, Webster, M.E, Mekel, M.J. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The development of monocyclic pyrazolone based cytokine synthesis inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1Y9U
 
 | |
6CKD
 
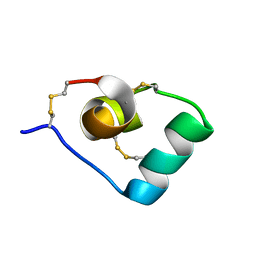 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p1 | | Descriptor: | OspTx2a-p1 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
8TEE
 
 | |
8TEC
 
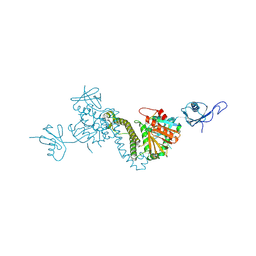 | |
1ZHC
 
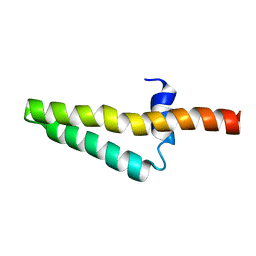 | |
1ZC4
 
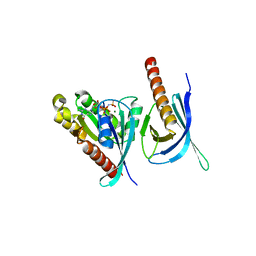 | | Crystal structure of the Ral-binding domain of Exo84 in complex with the active RalA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-A, ... | | Authors: | Jin, R, Junutula, J.R, Matern, H.T, Ervin, K.E, Scheller, R.H, Brunger, A.T. | | Deposit date: | 2005-04-10 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase.
Embo J., 24, 2005
|
|
4ATW
 
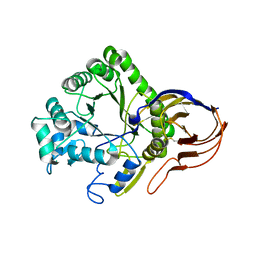 | | The crystal structure of Arabinofuranosidase | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE DOMAIN PROTEIN | | Authors: | Dumbrepatil, A, Song, H.-N, Jung, T.-Y, Kim, T.-J, Woo, E.-J. | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Alpha-L-Arabinofuranosidase from Thermotoga Maritima Reveals Characteristics for Thermostability and Substrate Specificity.
J.Microbiol.Biotech., 22, 2012
|
|
5WI3
 
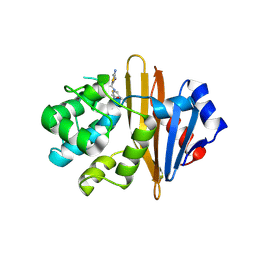 | | Structure of Acinetobacter baumannii carbapenemase OXA-239 K82D bound to cefotaxime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Harper, T.M, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2017-07-18 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239.
Biochem. J., 475, 2018
|
|
2X6R
 
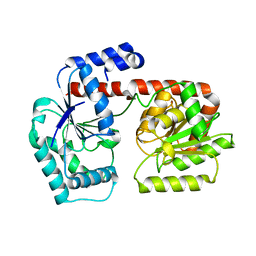 | | Crystal structure of trehalose synthase TreT from P.horikoshi produced by soaking in trehalose | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lim, M.-Y, Lee, S.-B, Woo, E.-J. | | Deposit date: | 2010-02-19 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2WSK
 
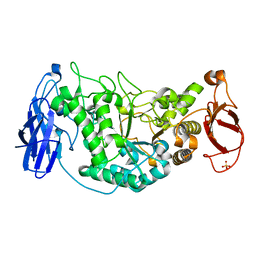 | | Crystal structure of Glycogen Debranching Enzyme GlgX from Escherichia coli K-12 | | Descriptor: | GLYCOGEN DEBRANCHING ENZYME, SULFATE ION | | Authors: | Song, H.-N, Park, J.-T, Jung, T.-Y, Park, K.-H, Woo, E.-J. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Rationale for the Short Branched Substrate Specificity of the Glycogen Debranching Enzyme Glgx.
Proteins, 78, 2010
|
|
2WTX
 
 | | Insight into the mechanism of enzymatic glycosyltransfer with retention through the synthesis and analysis of bisubstrate glycomimetics of trehalose-6-phosphate synthase | | Descriptor: | 1,2-ETHANEDIOL, ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE [UDP-FORMING], ... | | Authors: | Errey, J.C, Lee, S.S, Gibson, R.P, Martinez-Fleites, C, Barry, C.S, Jung, P.M.J, OSullivan, A, Davis, B.G, Davies, G.J. | | Deposit date: | 2009-09-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight Into Enzymatic Glycosyl Transfer with Retention of Configuration Through Analysis of Glycomimetic Inhibitors.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8BLR
 
 | | G13D mutant of KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation | | Descriptor: | GTPase KRas, N-terminally processed, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Moche, M, Jungholm, O, Strandback, E, Ampah-Korsah, H, Nyman, T, Orwar, O. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4 A crystal structure of K-Ras4A mutant G13D in open conformation
To Be Published
|
|
2X2G
 
 | | CRYSTALLOGRAPHIC BINDING STUDIES WITH AN ENGINEERED MONOMERIC VARIANT OF TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1U
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1S
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8PEE
 
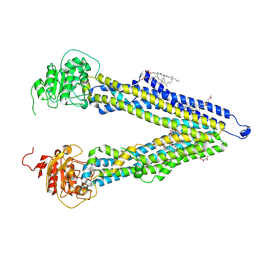 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
1YBY
 
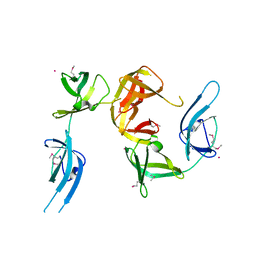 | | Conserved hypothetical protein Cth-95 from Clostridium thermocellum | | Descriptor: | Translation elongation factor P, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Zhou, W, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved hypothetical protein Cth-95 from Clostridium thermocellum
To be published
|
|
4AEF
 
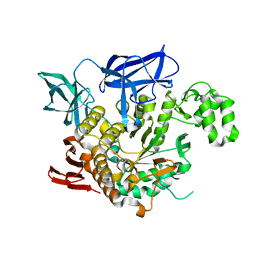 | | THE CRYSTAL STRUCTURE OF THERMOSTABLE AMYLASE FROM THE PYROCOCCUS | | Descriptor: | NEOPULLULANASE (ALPHA-AMYLASE II) | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Yang, S.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Biochim.Biophys.Acta, 1834, 2013
|
|
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
5WVR
 
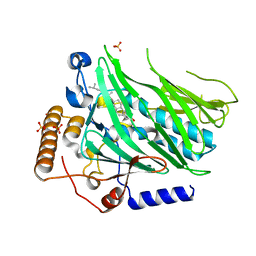 | | Crystal structure of Osh1 ORD domain in complex with cholesterol | | Descriptor: | CHOLESTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-12-28 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5WIB
 
 | | Structure of Acinetobacter baumannii carbapenemase OXA-239 K82D bound to imipenem | | Descriptor: | Imipenem, OXA-239 | | Authors: | Harper, T.M, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2017-07-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239.
Biochem. J., 475, 2018
|
|
