5BUR
 
 | | O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, in Complex with ATP and Magnesium Ion | | Descriptor: | 1,2-ETHANEDIOL, 2-succinylbenzoate--CoA ligase, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
3KFM
 
 | |
1KI2
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
4LZ8
 
 | |
5BUS
 
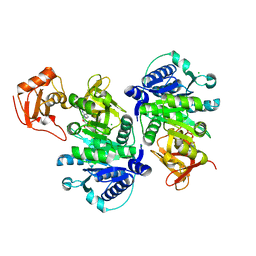 | | O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, in complex with AMP | | Descriptor: | 2-succinylbenzoate--CoA ligase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
3IT2
 
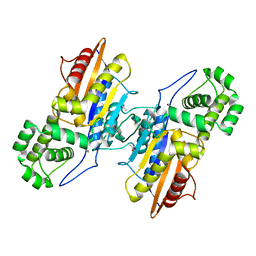 | | Crystal structure of ligand-free Francisella tularensis histidine acid phosphatase | | Descriptor: | ACETATE ION, Acid phosphatase | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3NY3
 
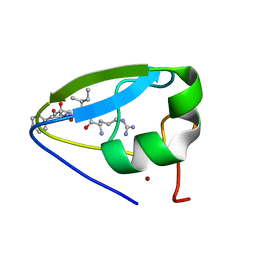 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2ERZ
 
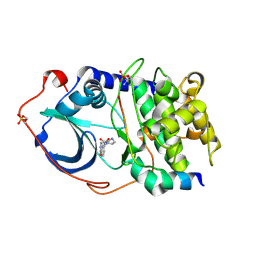 | | Crystal Structure of c-AMP Dependent Kinase (PKA) bound to hydroxyfasudil | | Descriptor: | 1-(1-HYDROXY-5-ISOQUINOLINESULFONYL)HOMOPIPERAZINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
2P1V
 
 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-propoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[4-HYDROXY-3-(5,5,8,8-TETRAMETHYL-3-PROPOXY-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P1T
 
 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-methoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[4-HYDROXY-3-(3-METHOXY-5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RTB
 
 | | APOSTREPTAVIDIN, PH 3.32, SPACE GROUP I222 | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTH
 
 | | STREPTAVIDIN-GLYCOLURIL COMPLEX, PH 2.50, SPACE GROUP I222 | | Descriptor: | ACETATE ION, GLYCOLURIL, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTP
 
 | |
2RTE
 
 | | STREPTAVIDIN-BIOTIN COMPLEX, PH 1.90, SPACE GROUP I222 | | Descriptor: | BIOTIN, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTF
 
 | | STREPTAVIDIN-BIOTIN COMPLEX, PH 2.00, SPACE GROUP I222 | | Descriptor: | BIOTIN, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTM
 
 | |
2RTJ
 
 | | STREPTAVIDIN-GLYCOLURIL COMPLEX, PH 2.50, SPACE GROUP I4122 | | Descriptor: | FORMIC ACID, GLYCOLURIL, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
1IT3
 
 | | Hagfish CO ligand hemoglobin | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin | | Authors: | Mito, M, Chong, K.T, Park, S.-Y, Tame, J.R. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of deoxy- and carbonmonoxyhemoglobin F1 from the hagfish Eptatretus burgeri
J.Biol.Chem., 277, 2002
|
|
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
6H41
 
 | |
5ZYI
 
 | | Crystal structure of CERT START domain in complex with compound E16 | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
1JPZ
 
 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
5ZYJ
 
 | | Crystal structure of CERT START domain in complex with compound E16A | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
3LBK
 
 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
5XGF
 
 | |
