2JBV
 
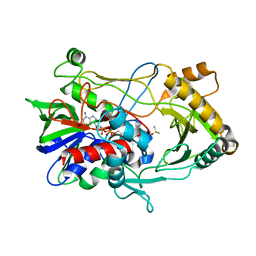 | | Crystal structure of choline oxidase reveals insights into the catalytic mechanism | | Descriptor: | CHOLINE OXIDASE, DIMETHYL SULFOXIDE, UNKNOWN ATOM OR ION, ... | | Authors: | Lountos, G.T, Fan, F, Gadda, G, Orville, A.M. | | Deposit date: | 2006-12-13 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of Glu312 in Binding and Positioning of the Substrate for the Hydride Transfer Reaction in Choline Oxidase.
Biochemistry, 47, 2008
|
|
1P3H
 
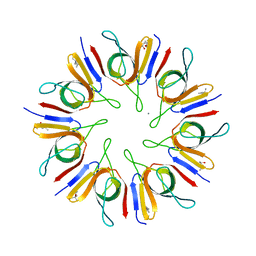 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
1RA1
 
 | |
1RA3
 
 | |
1RA2
 
 | |
1FQD
 
 | | CRYSTAL STRUCTURE OF MALTOTETRAITOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1RA8
 
 | |
1JP3
 
 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | Authors: | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | Deposit date: | 2001-07-31 | | Release date: | 2001-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
1P6H
 
 | | Rat neuronal NOS heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-2,4-L-DIAMINOBUTYRIC AMIDE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
7WGT
 
 | | X-ray structure of thermostabilized Drosophila dopamine transporter with GABA transporter1-like substitutions in the binding site, in complex with NO711. | | Descriptor: | 1-(2-{[(diphenylmethylidene)amino]oxy}ethyl)-1,2,5,6-tetrahydropyridine-3-carboxylic acid, Antibody fragment (9D5) heavy chain, Antibody fragment (9D5) light chain, ... | | Authors: | Joseph, D, Penmatsa, A. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Embo J., 41, 2022
|
|
2IGC
 
 | | Structure of Spin labeled T4 Lysozyme Mutant T115R1A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-09-22 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
1RX3
 
 | |
1KB9
 
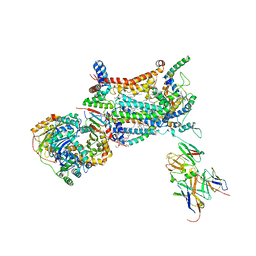 | | YEAST CYTOCHROME BC1 COMPLEX | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Lange, C, Nett, J.H, Trumpower, B.L, Hunte, C. | | Deposit date: | 2001-11-05 | | Release date: | 2002-09-18 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SPECIFIC ROLES OF PROTEIN-PHOSPHOLIPID INTERACTIONS IN THE YEAST CYTOCHROME BC1 COMPLEX STRUCTURE
EMBO J., 20, 2001
|
|
2IAH
 
 | | Crystal structure of the ferripyoverdine receptor of the outer membrane of Pseudomonas aeruginosa bound to ferripyoverdine. | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, FE (III) ION, Ferripyoverdine receptor, ... | | Authors: | Wirth, C, Pattus, F, Cobessi, D. | | Deposit date: | 2006-09-08 | | Release date: | 2007-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor.
J.Mol.Biol., 368, 2007
|
|
2IA6
 
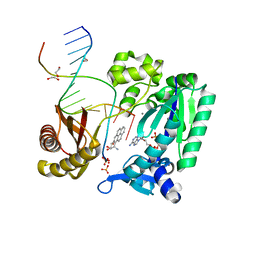 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*A)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7WJ7
 
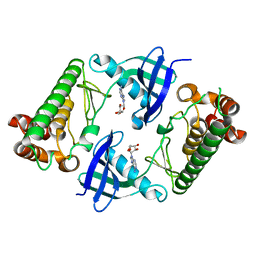 | |
1FOP
 
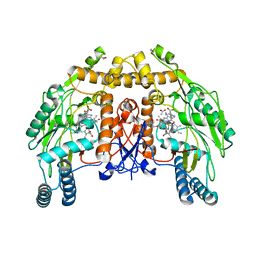 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH L-ARG AND NO(H4B-BOUND) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
1RX2
 
 | |
1FQA
 
 | | STRUCTURE OF MALTOTETRAITOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FIQ
 
 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7X5X
 
 | | Crystal structure of AtHPPD-YH20533 complex | | Descriptor: | 2-methylsulfonyl-N-(1-methyl-1,2,3,4-tetrazol-5-yl)-4-(trifluoromethyl)benzamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
2IWS
 
 | | Radicicol analogues bound to the ATP site of HSP90 | | Descriptor: | (5Z)-12-CHLORO-13,15-DIHYDROXY-4,7,8,9-TETRAHYDRO-2-BENZOXACYCLOTRIDECINE-1,10(3H,11H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
1PK9
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoroadenosine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PKE
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine and sulfate/phosphate | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PWL
 
 | | Crystal structure of human Aldose Reductase complexed with NADP and Minalrestat | | Descriptor: | 2[4-BROMO-2-FLUOROPHENYL)METHYL]-6-FLUOROSPIRO[ISOQUINOLINE-4-(1H),3'-PYRROLIDINE]-1,2',3,5'(2H)-TETRONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
