7PBI
 
 | |
7U4I
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | Descriptor: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7MQ2
 
 | |
8AH3
 
 | |
6KOB
 
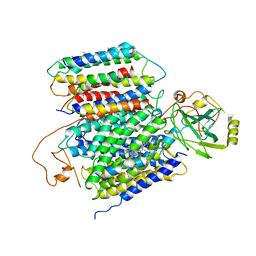 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ADR
 
 | |
5TON
 
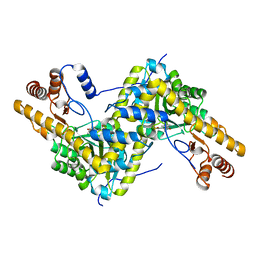 | | Crystal structure of AAT H143L mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
7P4Q
 
 | |
7MQ3
 
 | |
7P4P
 
 | |
5D2X
 
 | |
6LGK
 
 | |
5TPH
 
 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | Authors: | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
8AEJ
 
 | |
5JPZ
 
 | | Crystal structure of HAT domain of human Squamous Cell Carcinoma Antigen Recognized By T Cells 3, SART3 (TIP110) | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Grazette, A, Harper, S, Emsley, J, Layfield, R, Dreveny, I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | unpublished
To Be Published
|
|
5D2Y
 
 | |
5JQK
 
 | |
6KRM
 
 | |
8AJ0
 
 | | Mpro of SARS COV-2 in complex with the RK-90 inhibitor | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor RK-90
To Be Published
|
|
7TSX
 
 | | Structure of Enterobacter cloacae Cap2 bound to CdnD02 C-terminus, Apo state | | Descriptor: | Cap2, Cyclic AMP-AMP-GMP synthase | | Authors: | Ye, Q, Gu, Y, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
5JR6
 
 | |
5TOR
 
 | | Crystal structure of AAT D222T mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5D3E
 
 | |
7P6V
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH compound 3ag | | Descriptor: | Bromodomain-containing protein 4, ethyl 2-(2-(4-azido-N-((2-(1,5-dimethyl-6-oxo-1,6-dihydropyridin-3-yl)-1-((tetrahydro-2H-pyran-4-yl)methyl)-1H-benzo[d]imidazol-6-yl)methyl)-2,3,5,6-tetrafluorobenzamido)acetamido)acetate | | Authors: | Chung, C. | | Deposit date: | 2021-07-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | One-Step Synthesis of Photoaffinity Probes for Live-Cell MS-Based Proteomics.
Chemistry, 27, 2021
|
|
7MQ1
 
 | | C9A Streptococcus pneumoniae CstR in the reduced state, space group C2 | | Descriptor: | CHLORIDE ION, Copper-sensing transcriptional repressor csoR, GLYCEROL, ... | | Authors: | Fakhoury, J.N, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional asymmetry and chemical reactivity of CsoR family persulfide sensors.
Nucleic Acids Res., 49, 2021
|
|
