5ZRF
 
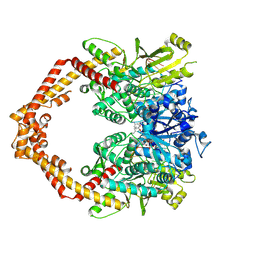 | | Crystal structure of human topoisomerase II beta in complex with 5-iodouridine-containing-DNA and etoposide in space group p21 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Chen, S.F, Wang, Y.R, Wu, C.C, Chan, N.L. | | Deposit date: | 2018-04-24 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the gating of DNA passage by the topoisomerase II DNA-gate.
Nat Commun, 9, 2018
|
|
3PJS
 
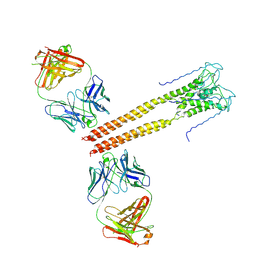 | | Mechanism of Activation Gating in the Full-Length KcsA K+ Channel | | Descriptor: | FAB heavy chain, FAB light chain, Voltage-gated potassium channel | | Authors: | Uysal, S, Cuello, L.G, Kossiakoff, A, Perozo, E. | | Deposit date: | 2010-11-10 | | Release date: | 2011-07-06 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of activation gating in the full-length KcsA K+ channel.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PKJ
 
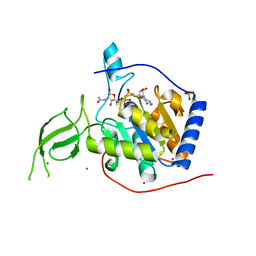 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
6A2J
 
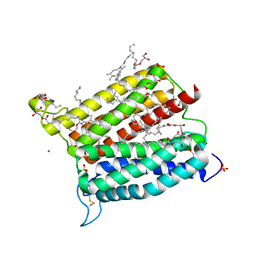 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-06-12 | | Release date: | 2018-11-21 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A7A
 
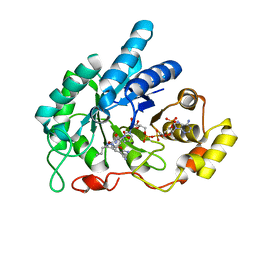 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
3PPH
 
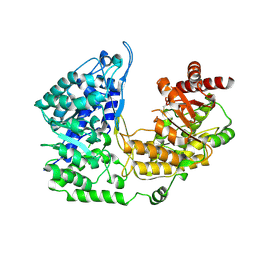 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, threonine variant | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
4LBN
 
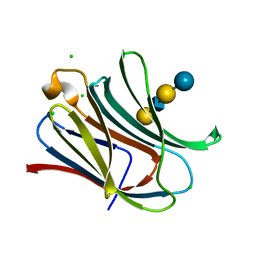 | | Crystal structure of Human galectin-3 CRD in complex with LNnT | | Descriptor: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2013-06-20 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Galectin-3 interactions with glycosphingolipids.
J.Mol.Biol., 426, 2014
|
|
3PUU
 
 | |
4L5Z
 
 | |
4L65
 
 | | Crystal structure of the Candida albicans Methionine Synthase in complex with 5-Methyl-Tetrahydrofolate and Methionine | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, METHIONINE, ... | | Authors: | Ubhi, D, Robertus, J.D. | | Deposit date: | 2013-06-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of a fungal methionine synthase with substrates and inhibitors.
J.Mol.Biol., 426, 2014
|
|
3RYB
 
 | | Lactococcal OppA complexed with SLSQSLSQS | | Descriptor: | Oligopeptide, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Importance of a Hydrophobic Pocket for Peptide Binding in Lactococcal OppA.
J.Bacteriol., 193, 2011
|
|
3RYC
 
 | | Tubulin: RB3 stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nawrotek, A, Knossow, M, Gigant, B. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Determinants That Govern Microtubule Assembly from the Atomic Structure of GTP-Tubulin.
J.Mol.Biol., 412, 2011
|
|
3S33
 
 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 10s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
3S3C
 
 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 360s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
3S3X
 
 | | Structure of chicken acid-sensing ion channel 1 AT 3.0 A resolution in complex with psalmotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, neuronal, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
4KT0
 
 | | Crystal structure of a virus like photosystem I from the cyanobacterium Synechocystis PCC 6803 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Mazor, Y, Nataf, D, Toporik, H, Nelson, N. | | Deposit date: | 2013-05-19 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803.
Elife, 3, 2014
|
|
3SBP
 
 | | Pseudomonas stutzeri nitrous oxide reductase, P1 crystal form | | Descriptor: | CALCIUM ION, CHLORIDE ION, DINUCLEAR COPPER ION, ... | | Authors: | Pomowski, A, Zumft, W.G, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase.
Nature, 477, 2011
|
|
3RYA
 
 | | Lactococcal OppA complexed with SLSQLSSQS | | Descriptor: | Oligopeptide, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Importance of a Hydrophobic Pocket for Peptide Binding in Lactococcal OppA.
J.Bacteriol., 193, 2011
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
4KCR
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-(3-(((3-fluorophenethyl)amino)methyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
3QAZ
 
 | | IL-2 mutant D10 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, Interleukin-2, ... | | Authors: | Levin, A.M, Bates, D.L, Ring, A.M, Lin, J.T, Su, L, Krieg, C, Bowman, G.R, Novick, P, Pande, V.S, Khort, H.E, Boyman, O, Gathman, C.G, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'
Nature, 484, 2012
|
|
4KCH
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with N,N'-([1,1'-biphenyl]-3,3'-diyl)bis(thiophene-2-carboximidamide) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N,N'-biphenyl-3,3'-diyldithiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
3QGP
 
 | |
3QI4
 
 | | Crystal structure of PDE9A(Q453E) in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
4KKO
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-((4-methoxy-6-(2-morpholinoethoxy)-1,3,5-triazin-2-yl)amino)-2-((3-methylbut-2-en-1-yl)oxy)benzonitrile (JLJ513), a non-nucleoside inhibitor | | Descriptor: | 4-({4-methoxy-6-[2-(morpholin-4-yl)ethoxy]-1,3,5-triazin-2-yl}amino)-2-(3-methylbutoxy)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Extension into the entrance channel of HIV-1 reverse transcriptase-Crystallography and enhanced solubility.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
