6JUU
 
 | | Crystal structure of ZAK in complex with compound 6r | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]naphthalene-1-sulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
2YVW
 
 | |
3OKS
 
 | |
6TP4
 
 | | Crystal structure of the Orexin-1 receptor in complex with ACT-462206 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2~{S})-~{N}-(3,5-dimethylphenyl)-1-(4-methoxyphenyl)sulfonyl-pyrrolidine-2-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5Z5I
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose and D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose, ... | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
4NPU
 
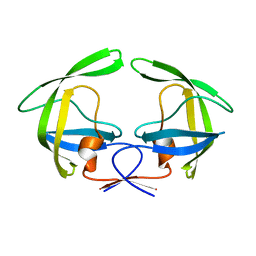 | |
4E4U
 
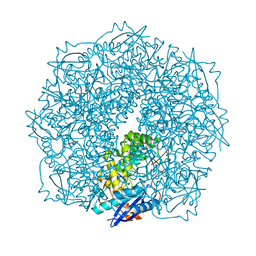 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200780) from Burkholderia SAR-1 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Mandalate racemase/muconate lactonizing enzyme | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-13 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Burkholderia SAR-1
to be published
|
|
1KRU
 
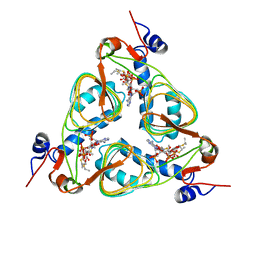 | |
1TZD
 
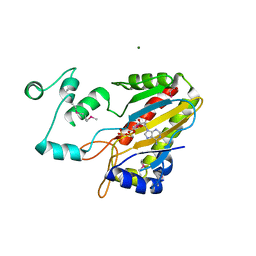 | | CRYSTAL STRUCTURE OF THE CATALYTIC CORE OF INOSITOL 1,4,5-TRISPHOSPHATE 3-KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MAGNESIUM ION | | Authors: | Miller, G.J, Hurley, J.H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase
Mol.Cell, 15, 2004
|
|
6TP6
 
 | | Crystal structure of the Orexin-1 receptor in complex with filorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CHLORIDE ION, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
3DN3
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
3H3K
 
 | |
3GNR
 
 | | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase with covalently bound 2-deoxy-2-fluoroglucoside to the catalytic nucleophile E396 | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, GLYCEROL, Os03g0212800 protein | | Authors: | Seshadri, S, Akiyama, T, Opassiri, R, Kuaprasert, B, Cairns, J.R.K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and enzymatic characterization of Os3BGlu6, a rice beta-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.
Plant Physiol., 151, 2009
|
|
3TUT
 
 | | Crystal structure of RtcA.ATP binary complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2011-09-18 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of RNA 3'-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3H5S
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor | | Descriptor: | (5S)-5-tert-butyl-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-3-[8-(methylsulfonyl)-1,1-dioxido-6,7,8,9-tetrahydroisothiazolo[4,5-h]isoquinolin-3-yl]-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2R5T
 
 | | Crystal Structure of Inactive Serum and Glucocorticoid- Regulated Kinase 1 in Complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Zhao, B, Lehr, R, Smallwood, A.M, Ho, T.F, Maley, K, Randall, T, Head, M.S, Koretke, K.K, Schnackenberg, C.G. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the kinase domain of serum and glucocorticoid-regulated kinase 1 in complex with AMP PNP.
Protein Sci., 16, 2007
|
|
4IJN
 
 | |
5Z1T
 
 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(6-hydroxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
3GJW
 
 | | PARP complexed with A968427 | | Descriptor: | 7-(pyrrolidin-1-ylmethyl)pyrrolo[1,2-a]quinoxalin-4(5H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and SAR of novel tricyclic quinoxalinone inhibitors of poly(ADP-ribose)polymerase-1 (PARP-1)
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4YAB
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 1-methyl-5-(2-methyl-1 3-thiazol-4-yl)-2 3-dihydro-1H-indol-2-one (1) | | Descriptor: | 1-methyl-5-(2-methyl-1,3-thiazol-4-yl)-1,3-dihydro-2H-indol-2-one, SULFATE ION, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
6TOS
 
 | | Crystal structure of the Orexin-1 receptor in complex with GSK1059865 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7WVB
 
 | | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1 | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state
To Be Published
|
|
3EPB
 
 | | Human AdoMetDC E256Q mutant complexed with putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
7BVQ
 
 | | Structure of human beta1 adrenergic receptor bound to carazolol | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
8WKU
 
 | | Complex structure of MjHKU4r-CoV-1 spike RBD bound to human CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form, MjHKU4r-CoV-1 spike RBD, ... | | Authors: | Zhao, Z.N, Chai, Y, Gao, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for receptor recognition and broad host tropism for merbecovirus MjHKU4r-CoV-1.
Embo Rep., 25, 2024
|
|
