7T9L
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
5I23
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF022 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-{[(1S,2R,3S,4S,5R,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)cyclohexyl]amino}octyl)triaza-1,2-dien-2-ium, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
8HOD
 
 | | ScRIPK MUTANT-S253A, T254A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Hypothetical protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
3M09
 
 | | F98Y TMP-resistant Dihydrofolate Reductase from Staphylococcus aureus with inhibitor RAB1 | | Descriptor: | 5-(3,4-dimethoxy-5-{(1E)-3-oxo-3-[(1S)-1-propylphthalazin-2(1H)-yl]prop-1-en-1-yl}benzyl)pyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Inhibition of Antibiotic-Resistant Staphylococcus aureus by the Broad-Spectrum Dihydrofolate Reductase Inhibitor RAB1.
Antimicrob.Agents Chemother., 54, 2010
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3M3Y
 
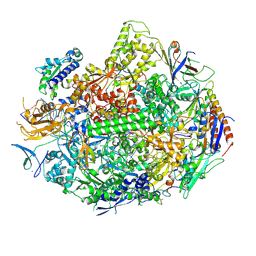 | | RNA polymerase II elongation complex C | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8H3Q
 
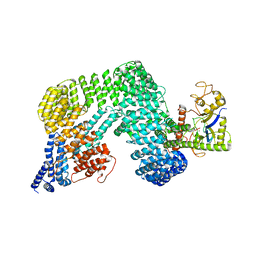 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | Descriptor: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6HRG
 
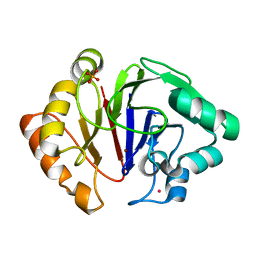 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
7T71
 
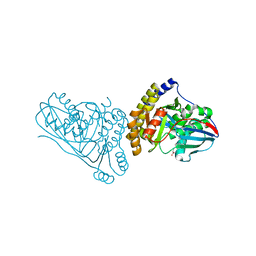 | | Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus | | Descriptor: | Mevalonate 3,5-bisphosphate decarboxylase, OLEIC ACID | | Authors: | Vinokur, J.M, Sawaya, M.R, Cascio, D, Collazo, M, Bowie, J.U. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways.
J.Biol.Chem., 298, 2022
|
|
8H35
 
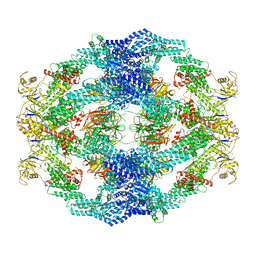 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.41 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4BX9
 
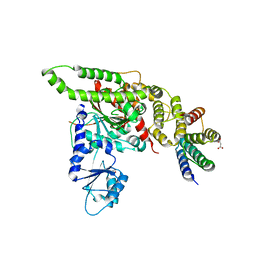 | | Human Vps33A in complex with a fragment of human Vps16 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FORMIC ACID, MALONIC ACID, ... | | Authors: | Graham, S.C, Wartosch, L, Gray, S.R, Scourfield, E.J, Deane, J.E, Luzio, J.P, Owen, D.J. | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Vps33A recruitment to the human HOPS complex by Vps16.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
2DH2
 
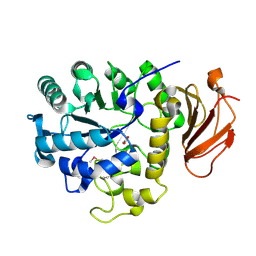 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ACETATE ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
4C9Y
 
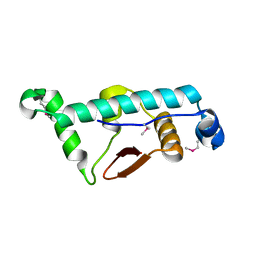 | | Structural Basis for the microtubule binding of the human kinetochore Ska complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1 | | Authors: | Abad, M, Medina, B, Santamaria, A, Zou, J, Plasberg-Hill, C, Madhumalar, A, Jayachandran, U, Redli, P.M, Rappsilber, J, Nigg, E.A, Jeyaprakash, A.A. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for Microtubule Recognition by the Human Kinetochore Ska Complex.
Nat.Commun., 5, 2014
|
|
7T9J
 
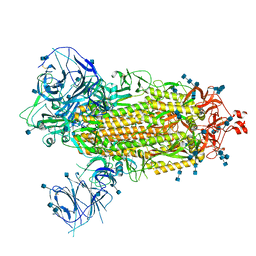 | | Cryo-EM structure of the SARS-CoV-2 Omicron spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7T9K
 
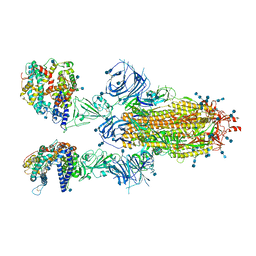 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
5I6U
 
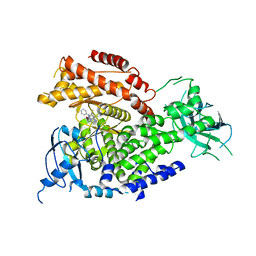 | | The crystal structure of PI3Kdelta with compound 32 | | Descriptor: | 2-[(1S)-1-({6-amino-5-[(1H-pyrazol-4-yl)ethynyl]pyrimidin-4-yl}amino)ethyl]-5-chloro-3-phenylquinazolin-4(3H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2016-02-16 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | The crystal structure of PI3Kdelta with compound 32
To Be Published
|
|
8PW7
 
 | | A respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H33
 
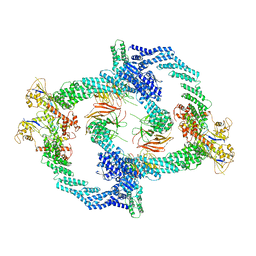 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.86 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4BS2
 
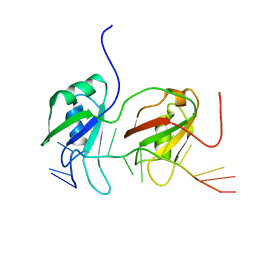 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
7SQM
 
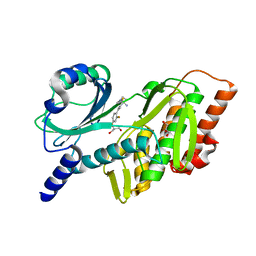 | | Discovery and Preclinical Pharmacology of INE963, A Potent and Fast-Acting Blood-Stage Antimalarial with a High Barrier to Resistance and Potential for Single-Dose Cure in Uncomplicated Malaria | | Descriptor: | 1-[(4S)-5-(2,4-difluorophenyl)imidazo[2,1-b][1,3,4]thiadiazol-2-yl]-4-methylpiperidin-4-amine, GLYCEROL, Serine/threonine-protein kinase haspin | | Authors: | Shu, W, Yokokawa, F. | | Deposit date: | 2021-11-05 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Failure of artesunate-mefloquine combination therapy for uncompli-cated Plasmodium falciparum malaria in southern Cambodia.
Malar. J., 2009, 8, 10, 2009
|
|
3MBG
 
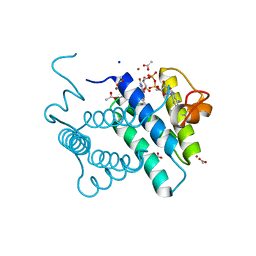 | | Crystal Structure of Human Augmenter of Liver Regeneration (ALR) | | Descriptor: | ACETATE ION, FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dong, M, Schaefer, S, Daithankar, V.N, Thorpe, C, Bahnson, B.J. | | Deposit date: | 2010-03-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy .
Biochemistry, 49, 2010
|
|
3M6M
 
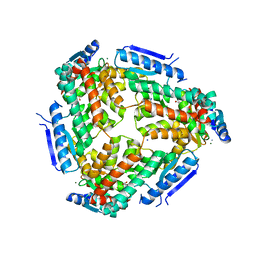 | | Crystal structure of RpfF complexed with REC domain of RpfC | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Cheng, Z, Lim, S.C, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
3MBT
 
 | | Structure of monomeric Blc from E. coli | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2010-03-26 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses reveal a monomeric state of the bacterial lipocalin Blc.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ADG
 
 | |
