5X8A
 
 | | Crystal structure of ATP bound thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5X8C
 
 | | AMPPCP and TMP bound crystal structure of thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5X8J
 
 | |
5X99
 
 | |
4LAJ
 
 | | Crystal structure of HIV-1 YU2 envelope gp120 glycoprotein in complex with CD4-mimetic miniprotein, M48U1, and llama single-domain, broadly neutralizing, co-receptor binding site antibody, JM4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Heavy Chain-Only IgG2b Llama Antibody Effects Near-Pan HIV-1 Neutralization by Recognizing a CD4-Induced Epitope That Includes Elements of Coreceptor- and CD4-Binding Sites.
J.Virol., 87, 2013
|
|
5KNN
 
 | |
3KFD
 
 | |
1UC6
 
 | | Solution Structure of the Carboxyl Terminal Domain of the Ciliary Neurotrophic Factor Receptor | | Descriptor: | Ciliary Neurotrophic Factor Receptor alpha | | Authors: | Man, D, He, W, Sze, K.H, Ke, G, Smith, D.K, Ip, N.Y, Zhu, G. | | Deposit date: | 2003-04-08 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of the ciliary neurotrophic factor (CNTF) receptor and ligand free associations among components of the CNTF receptor complex
J.Biol.Chem., 278, 2003
|
|
1WKW
 
 | | Crystal structure of the ternary complex of eIF4E-m7GpppA-4EBP1 peptide | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Matsushita, Y, Fujisaki, H, Shen, X, Miyagawa, H, Kitamura, K, Miura, K, Ishida, T. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods
Biochim.Biophys.Acta, 1753, 2005
|
|
7UUT
 
 | | Ternary complex crystal structure of secondary alcohol dehydrogenases from the Thermoanaerobacter ethanolicus mutants C295A and I86A provides better understanding of catalytic mechanism | | Descriptor: | (2R)-pentan-2-ol, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Dinh, T, Phillips, R, Rahn, K. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ternary complexes of thermophilic secondary alcohol dehydrogenase from Thermoanaerobacter pseudoethanolicus reveal the dynamics of ligand exchange and the proton relay network.
Proteins, 90, 2022
|
|
5ZAX
 
 | | Crystal structure of thymidylate kinase in complex with ADP, TDP and TMP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2018
|
|
5ZB4
 
 | | Crystal structure of thymidylate kinase in complex with ADP and TMP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2018
|
|
5ZB0
 
 | | Crystal structure of thymidylate kinase in complex with ADP and TDP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2018
|
|
6LVE
 
 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3RJJ
 
 | | Ternary complex crystal structure of DNA Polymerase Beta with template 8odG provides insight into mutagenic lesion bypass | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*(8OG)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RJK
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG:dC base pair at primer terminus and dG:dCMP(CF2)PP in the active site | | Descriptor: | 2'-deoxy-5'-O-[(S)-{difluoro[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*GP*(8OG)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RJI
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position paired with non-hydrolyzable dCTP analog (dCMP(CF2)PP) | | Descriptor: | 2'-deoxy-5'-O-[(S)-{difluoro[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RJH
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing (syn)8odG:dA at primer terminus and dG:dCMP(CF2)PPin the active site | | Descriptor: | 2'-deoxy-5'-O-[(S)-{difluoro[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*GP*(8OG)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RJF
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing (syn)8odG at template position paired with non-hydrolyzable dATP analog (dApCPP) | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RJE
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DMO
 
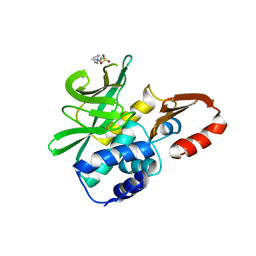 | | Crystal structure of the (BACCR)NAT3 arylamine N-acetyltransferase from Bacillus cereus reveals a unique Cys-His-Glu catalytic triad | | Descriptor: | 3-(1-methylpiperidinium-1-yl)propane-1-sulfonate, N-hydroxyarylamine O-acetyltransferase | | Authors: | Kubiak, X, Li de la Sierra-Gallay, I, Haouz, A, Weber, P, Rodrigues-Lima, F. | | Deposit date: | 2012-02-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and Biochemical Characterization of an Active Arylamine N-Acetyltransferase Possessing a Non-canonical Cys-His-Glu Catalytic Triad.
J.Biol.Chem., 288, 2013
|
|
3M9D
 
 | |
3M91
 
 | |
7U6L
 
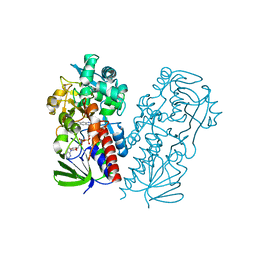 | | Pseudooxynicotine amine oxidase | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choudhary, V, Stull, F, Wu, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
