4JPC
 
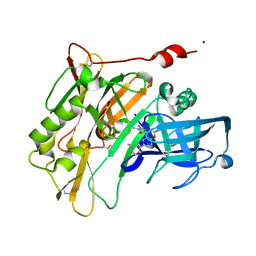 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | Descriptor: | 3-[(4R)-2'-amino-1',2,2-trimethyl-5'-oxo-1',2,3,5'-tetrahydrospiro[chromene-4,4'-imidazol]-6-yl]benzonitrile, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
3L59
 
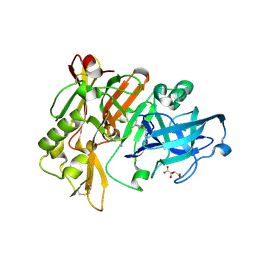 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
4MI9
 
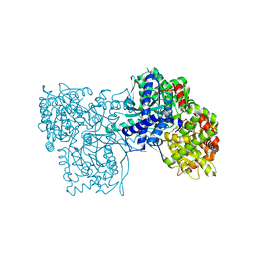 | | Crystal structure of Gpb in complex with SUGAR (N-[(3R)-3-(4-ETHYLPHENYL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) (S20) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(3R)-3-(4-ethylphenyl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
2POG
 
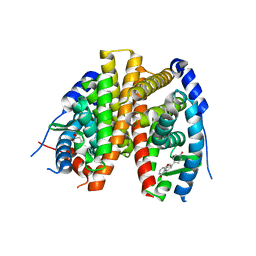 | | Benzopyrans as Selective Estrogen Receptor b Agonists (SERBAs). Part 2: Structure Activity Relationship Studies on the Benzopyran Scaffold. | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-9-OL, Estrogen receptor | | Authors: | Richardson, T.I, Norman, B.H, Lugar, C.W, Jones, S.A, Wang, Y, Durbin, J.D, Krishnan, V, Dodge, J.A. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 2: structure-activity relationship studies on the benzopyran scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2NO7
 
 | | C4S dCK variant of dCK in complex with L-dC+ADP | | Descriptor: | 4-AMINO-1-(2-DEOXY-BETA-L-ERYTHRO-PENTOFURANOSYL)PYRIMIDIN-2(1H)-ONE, ADENOSINE-5'-DIPHOSPHATE, deoxycytidine kinase | | Authors: | Sabini, E, Hazra, S, Konrad, M, Burley, S.K, Lavie, A. | | Deposit date: | 2006-10-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonenantioselectivity Property of Human Deoxycytidine Kinase Explained by Structures of the Enzyme in Complex with l- and d-Nucleosides.
J.Med.Chem., 50, 2007
|
|
4JP9
 
 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | Descriptor: | (4R)-2'-amino-6-(3-chlorophenyl)-1',2,2-trimethyl-2,3-dihydrospiro[chromene-4,4'-imidazol]-5'(1'H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
3OY0
 
 | | Human Carbonic Anhydrase II complexed with 1-(4-(4-(2-(ISOPROPYLSULFONYL)PHENYLAMINO)-1H-PYRROLO[2,3-B]PYRIDIN-6-YLAMINO)-3-METHOXYPHENYL)PIPERIDIN-4-OL | | Descriptor: | (2S)-2-tert-butyl-N-(4-sulfamoylphenyl)pentanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Aggarwal, M, McKenna, R. | | Deposit date: | 2010-09-22 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anticonvulsant 4-aminobenzenesulfonamide derivatives with branched-alkylamide moieties: X-ray crystallography and inhibition studies of human carbonic anhydrase isoforms I, II, VII, and XIV.
J.Med.Chem., 54, 2011
|
|
2NO0
 
 | | C4S dCK variant of dCK in complex with gemcitabine+ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, GEMCITABINE | | Authors: | Sabini, E, Hazra, S, Konrad, M, Burley, S.K, Lavie, A. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonenantioselectivity Property of Human Deoxycytidine Kinase Explained by Structures of the Enzyme in Complex with l- and d-Nucleosides.
J.Med.Chem., 50, 2007
|
|
2NO1
 
 | | C4S dCK variant of dCK in complex with D-dC+ADP | | Descriptor: | 2'-DEOXYCYTIDINE, ADENOSINE-5'-DIPHOSPHATE, deoxycytidine kinase | | Authors: | Sabini, E, Hazra, S, Konrad, M, Burley, S.K, Lavie, A. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Nonenantioselectivity Property of Human Deoxycytidine Kinase Explained by Structures of the Enzyme in Complex with l- and d-Nucleosides.
J.Med.Chem., 50, 2007
|
|
3A29
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor | | Descriptor: | 2-amino-4,5-dihydronaphtho[1,2-d][1,3]thiazol-8-yl dihydrogen phosphate, Fructose-1,6-bisphosphatase 1 | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR, and X-ray structure of tricyclic compounds as potent FBPase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3TTJ
 
 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3BR9
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | (2R)-2-({3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothia diazin-7-yl}oxy)propanamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BSC
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 5-hydroxy-4-(7-methoxy-1,1-dioxido-2H-1,2,4-benzothiadiazin-3-yl)-2-(3-methylbutyl)-6-phenylpyridazin-3(2H)-one, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BSA
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 2-({3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-(1,3-thiazol-5-yl)-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothia diazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2GEH
 
 | | N-Hydroxyurea, a versatile zinc binding function in the design of metalloenzyme inhibitors | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, N-HYDROXYUREA, ... | | Authors: | Temperini, C, Innocenti, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Hydroxyurea-A versatile zinc binding function in the design of metalloenzyme inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3LFN
 
 | | Crystal structure of CDK2 with SAR57, an aminoindazole type inhibitor | | Descriptor: | Cell division protein kinase 2, N-[6-(4-hydroxyphenyl)-5-phenyl-1H-indazol-3-yl]butanamide | | Authors: | Dreyer, M.K, Wendt, K.U, Schimanski-Breves, S, Loenze, P. | | Deposit date: | 2010-01-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Rational design of potent GSK3beta inhibitors with selectivity for Cdk1 and Cdk2.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3Q32
 
 | | Structure of Janus kinase 2 with a pyrrolotriazine inhibitor | | Descriptor: | 2-(2,6-difluoro-4-methoxyphenyl)-1-(4-{4-[(3-methyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}piperazin-1-yl)ethanone, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrrolo[1,2-f]triazines as JAK2 inhibitors: Achieving potency and selectivity for JAK2 over JAK3.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2QP6
 
 | |
3LAU
 
 | |
3DD8
 
 | | Carbonic anhydrase inhibitors. Interaction of the antitumor sulfamate EMD-486019 with twelve mammalian isoforms: kinetic and X-Ray crystallographic studies | | Descriptor: | 2-(cycloheptylmethyl)-1,1-dioxido-1-benzothiophen-6-yl sulfamate, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Innocenti, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-06-05 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of the antitumor sulfamate EMD 486019 with twelve mammalian carbonic anhydrase isoforms: Kinetic and X-ray crystallographic studies
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GOL
 
 | | HCV NS5b polymerase in complex with 1,5 benzodiazepine inhibitor (R)-11d | | Descriptor: | (11R)-10-acetyl-11-(2,4-dichlorophenyl)-6-hydroxy-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Nyanguile, O, De Bondt, H. | | Deposit date: | 2009-03-19 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 1,5-Benzodiazepine inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3LE6
 
 | | The structure of cyclin dependent kinase 2 (CKD2) with a pyrazolobenzodiazepine inhibitor | | Descriptor: | 5-(2-chlorophenyl)-3-methyl-7-nitropyrazolo[3,4-b][1,4]benzodiazepine, Cell division protein kinase 2 | | Authors: | Lukacs, C.M, Swain, A, Crowther, R.L, Kammlott, R.U, Liu, J.J. | | Deposit date: | 2010-01-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolobenzodiazepines: part I. Synthesis and SAR of a potent class of kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LY2
 
 | | Catalytic Domain of Human Phosphodiesterase 4B in Complex with A Coumarin-Based Inhibitor | | Descriptor: | 8-(cyclopentyloxy)-4-[(3,5-dichloropyridin-4-yl)amino]-7-methoxy-2H-chromen-2-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Shiau, A.K, Coyle, A.R, Hsien, J.H, Staszewski, L.M. | | Deposit date: | 2010-02-26 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Water-soluble PDE4 inhibitors for the treatment of dry eye.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4DK8
 
 | | Crystal structure of LXR ligand binding domain in complex with partial agonist 5 | | Descriptor: | ACETATE ION, CALCIUM ION, N-methyl-N-(4-{(1S)-2,2,2-trifluoro-1-hydroxy-1-[1-(2-methoxyethyl)-1H-pyrrol-2-yl]ethyl}phenyl)benzenesulfonamide, ... | | Authors: | Piper, D.E, Kopecky, D.J, Xu, H. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of a new binding mode for a series of liver X receptor agonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3PS6
 
 | |
