5L0V
 
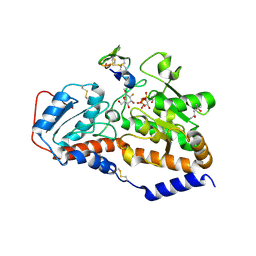 | | human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-beta-D-glucopyranose, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5HGO
 
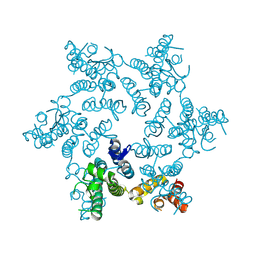 | | Hexameric HIV-1 CA R18G mutant | | Descriptor: | Capsid protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
3MDB
 
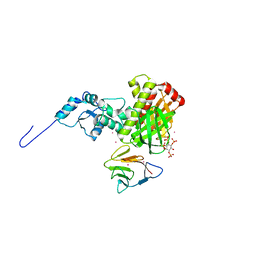 | | Crystal structure of the ternary complex of full length centaurin alpha-1, KIF13B FHA domain, and IP4 | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, Arf-GAP with dual PH domain-containing protein 1, Kinesin-like protein KIF13B, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Crystal structure of the ternary complex of full length centaurin alpha-1, KIF13B FHA domain, and IP4
to be published
|
|
4LQD
 
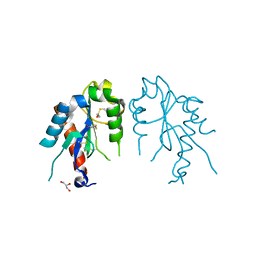 | | The crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry | | Descriptor: | GLYCEROL, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Snyder, G.A, Smith, P, Jiang, J, Xiao, T.S. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry.
J.Biol.Chem., 289, 2014
|
|
5T0R
 
 | |
5T0S
 
 | | Synaptotagmin 1 C2B domain, cadmium-bound | | Descriptor: | CADMIUM ION, SODIUM ION, Synaptotagmin-1 | | Authors: | Taylor, A.B, Hart, P.J, Igumenova, T.I. | | Deposit date: | 2016-08-16 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Non-Native Metal Ion Reveals the Role of Electrostatics in Synaptotagmin 1-Membrane Interactions.
Biochemistry, 56, 2017
|
|
1KY5
 
 | | D244E mutant S-Adenosylhomocysteine hydrolase refined with noncrystallographic restraints | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3'-OXO-ADENOSINE, S-adenosylhomocysteine hydrolase | | Authors: | Takata, Y, Takusagawa, F. | | Deposit date: | 2002-02-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic Mechanism of S-adenosylhomocysteine hydrolase. Site-directed
mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190.
J.Biol.Chem., 277, 2002
|
|
5L1Z
 
 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze-Gahmen, U, Hurley, J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
5FV7
 
 | | Human Fen1 in complex with an N-hydroxyurea compound | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, FLAP ENDONUCLEASE 1, MAGNESIUM ION | | Authors: | Exell, J.C, Thompson, M.J, Finger, L.D, Shaw, S.K, Abbott, W.M, McWhirter, C, Debreczeni, J.E, Jones, C.D, Nissink, J.W.M, Ward, T.A, Sioberg, C.W.L, Molina, D.M, Durant, S.T, Grasby, J.A. | | Deposit date: | 2016-02-03 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Cellular Active N-Hydroxyurea Fen1 Inhibitors Block Substrate Entry to the Active Site
Nat.Chem.Biol., 12, 2016
|
|
1POE
 
 | |
9GKE
 
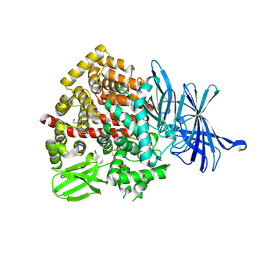 | | ERAP1 in complex with 1-[2-(2-oxo-5-phenyl-2,3-dihydro-1,3-benzothiazol-3-yl)acetamido]cyclohexane-1-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(2-oxidanylidene-5-phenyl-1,3-benzothiazol-3-yl)ethanoylamino]cyclohexane-1-carboxylic acid, Endoplasmic reticulum aminopeptidase 1, ... | | Authors: | Chung, C. | | Deposit date: | 2024-08-23 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Optimization of Potent and Selective Cyclohexyl Acid ERAP1 Inhibitors Using Structure- and Property-Based Drug Design.
Acs Med.Chem.Lett., 15, 2024
|
|
3MFK
 
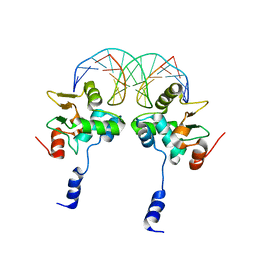 | |
6U2L
 
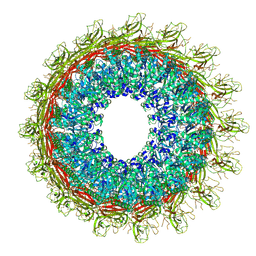 | | EM structure of MPEG-1 (L425K, beta conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6E4C
 
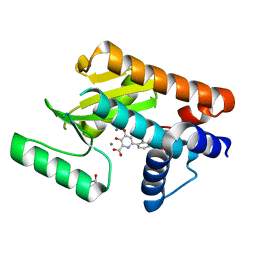 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-(2-methyl-4-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)phenyl)-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-[2-methyl-4-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)phenyl]-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
1L6N
 
 | |
6EQU
 
 | | X-Ray crystal structure of the human carbonic anhydrase II adduct with a membrane-impermeant inhibitor | | Descriptor: | 4-[2-(2,4,6-triphenylpyridin-1-ium-1-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human carbonic anhydrase II adduct with 1-(4-sulfamoylphenyl-ethyl)-2,4,6-triphenylpyridinium perchlorate, a membrane-impermeant, isoform selective inhibitor.
J Enzyme Inhib Med Chem, 33, 2018
|
|
7W33
 
 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | Descriptor: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
6ST8
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
4Z78
 
 | |
5EM8
 
 | |
5T6H
 
 | |
5MHF
 
 | | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase, ... | | Authors: | Hill, J.C, Caputo, A.T, Roversi, P, Zitzmann, N. | | Deposit date: | 2016-11-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
J.Med.Chem., 2020
|
|
6BSY
 
 | | HIV-1 Rev assembly domain (residues 1-69) | | Descriptor: | PHOSPHATE ION, Protein Rev | | Authors: | Watts, N.R, Eren, E, Zhuang, X, Wang, Y.X, Steven, A.C, Wingfield, P.T. | | Deposit date: | 2017-12-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A new HIV-1 Rev structure optimizes interaction with target RNA (RRE) for nuclear export.
J. Struct. Biol., 203, 2018
|
|
8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
