8JQB
 
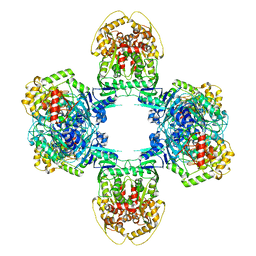 | | Structure of Gabija GajA-GajB 4:4 Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
6XDR
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 27 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
2E1W
 
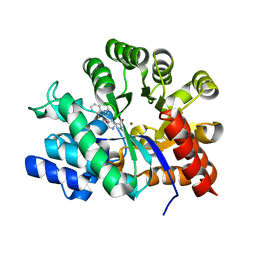 | | Crystal structure of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(1-NAPHTHYL)ETHYL]PROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design and Synthesis of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors
J.Med.Chem., 47, 2004
|
|
2WU3
 
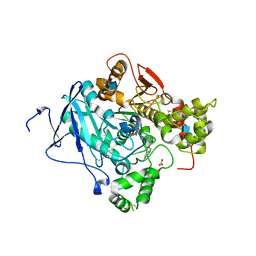 | | CRYSTAL STRUCTURE OF MOUSE ACETYLCHOLINESTERASE IN COMPLEX WITH FENAMIPHOS AND HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Hornberg, A, Artursson, E, Warme, R, Pang, Y.-P, Ekstrom, F. | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Oxime-Bound Fenamiphos-Acetylcholinesterases: Reactivation Involving Flipping of the His447 Ring to Form a Reactive Glu334-His447-Oxime Triad.
Biochem.Pharm., 79, 2010
|
|
2XI4
 
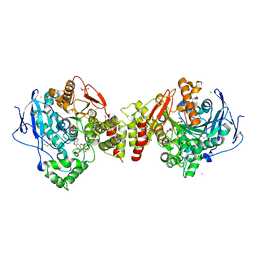 | | Torpedo californica Acetylcholinesterase in Complex with Aflatoxin B1 (Orthorhombic Space Group) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, AFLATOXIN B1, ... | | Authors: | Sanson, B, Colletier, J.P, Xu, Y, Lang, P.T, Jiang, H, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2010-06-28 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Backdoor Opening Mechanism in Acetylcholinesterase Based on X-Ray Crystallography and Molecular Dynamics Simulations.
Protein Sci., 20, 2011
|
|
2XBG
 
 | |
6XZB
 
 | | E. coli 70S ribosome in complex with dirithromycin, fMet-Phe-tRNA(Phe) and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
6XD0
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-434 | | Descriptor: | 2-[({2-[(2,6-dichlorophenyl)amino]phenyl}acetyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
6XJV
 
 | | MCU holocomplex in High-calcium state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
2E5C
 
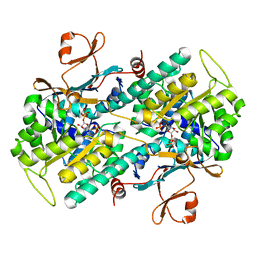 | | Crystal structure of Human NMPRTase complexed with 5'-phosphoribosyl-1'-pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2ERJ
 
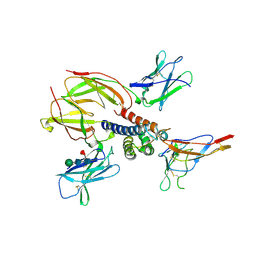 | | Crystal structure of the heterotrimeric interleukin-2 receptor in complex with interleukin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debler, E.W, Stauber, D.J, Wilson, I.A. | | Deposit date: | 2005-10-25 | | Release date: | 2006-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the IL-2 signaling complex: Paradigm for a heterotrimeric cytokine receptor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8KCB
 
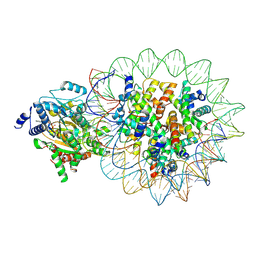 | | Complex of DDM1-nucleosome(H2A) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
8KCC
 
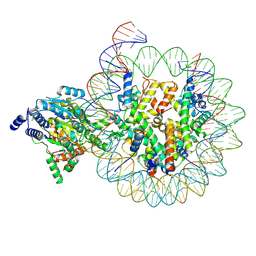 | | Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
2VZI
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD4 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin,Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
2ETM
 
 | |
2EYZ
 
 | | CT10-Regulated Kinase isoform II | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8OFB
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch, hexagonal form | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Reverse gyrase, ... | | Authors: | Klostermeier, D, Rasche, R, Mhaindarkar, V, Kummel, D, Rudolph, M.G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2YFQ
 
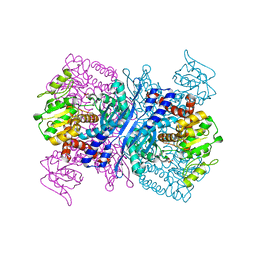 | | Crystal structure of Glutamate dehydrogenase from Peptoniphilus asaccharolyticus | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Oliveira, T, Panjikar, S, Carrigan, J.B, Sharkey, M.A, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-07 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of Nad-Dependent Peptoniphilus Asaccharolyticus Glutamate Dehydrogenase Reveals Determinants of Cofactor Specificity.
J.Struct.Biol., 177, 2012
|
|
8OIP
 
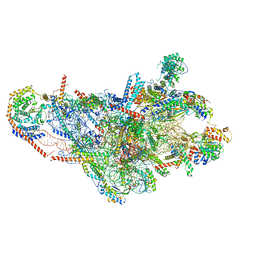 | | 28S mammalian mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S15, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
2FKX
 
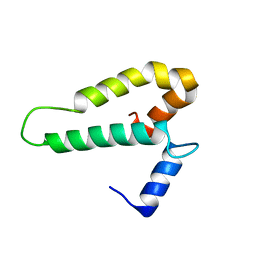 | |
6YN0
 
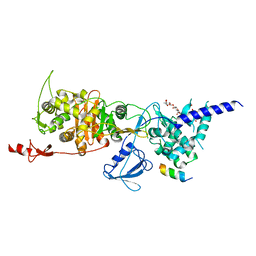 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
8OIS
 
 | | 28S human mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIT
 
 | | 39S human mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 16S rRNA, 39S ribosomal protein L1, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIQ
 
 | | 39S mammalian mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 16S rRNA, 39S ribosomal protein L1, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
2WKQ
 
 | | Structure of a photoactivatable Rac1 containing the Lov2 C450A Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
