1VFB
 
 | |
3V8E
 
 | | Crystal structure of the yeast nicotinamidase Pnc1p bound to the inhibitor nicotinaldehyde | | Descriptor: | MAGNESIUM ION, Nicotinamidase, ZINC ION | | Authors: | Hoadley, K.A, Smith, B.C, Denu, J.M, Keck, J.L. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Kinetic Isotope Effect Studies of Nicotinamidase (Pnc1) from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
4ZS6
 
 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
4ZT0
 
 | |
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
6RGJ
 
 | | Photorhabdus asymbiotica lectin PHL in complex with D-glucose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lectin PHL, ... | | Authors: | Houser, J, Fujdiarova, E, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
5WQ6
 
 | | Crystal Structure of hMNDA-PYD with MBP tag | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MBP tagged hMNDA-PYD, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
3VBQ
 
 | |
4V0K
 
 | |
3VIQ
 
 | | Crystal structure of Swi5-Sfr1 complex from fission yeast | | Descriptor: | GLYCEROL, Mating-type switching protein swi5, NITRATE ION, ... | | Authors: | Kuwabara, N, Murayama, Y, Hashimoto, H, Kokabu, Y, Ikeguchi, M, Sato, M, Mayanagi, K, Tsutsui, Y, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
4V1P
 
 | | BTN3 Structure | | Descriptor: | BUTYROPHILIN SUBFAMILY 3 MEMBER A1 | | Authors: | James, L.C. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Activation of Human Gammadelta T Cells by Cytosolic Interactions of Btn3A1 with Soluble Phosphoantigens and the Cytoskeletal Adaptor Periplakin.
J.Immunol., 194, 2015
|
|
3VM0
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - NO2 complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
4ZS7
 
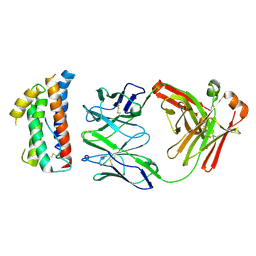 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
5WSF
 
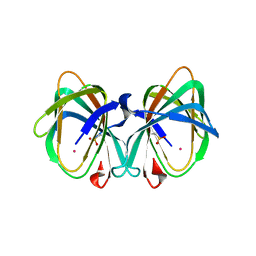 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
1R5D
 
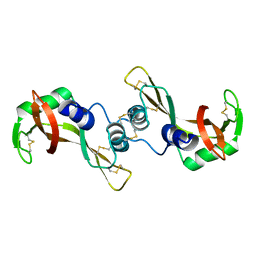 | | X-ray structure of bovine seminal ribonuclease swapping dimer from a new crystal form | | Descriptor: | Ribonuclease, seminal | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Population shift vs induced fit: The case of bovine seminal ribonuclease swapping dimer
Biopolymers, 73, 2004
|
|
4UX4
 
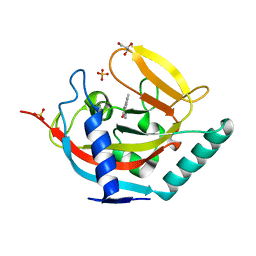 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | Descriptor: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
3VOK
 
 | | X-ray Crystal Structure of Wild Type HrtR in the Apo Form with the Target DNA. | | Descriptor: | 5'-D(*AP*TP*GP*AP*CP*AP*CP*TP*GP*TP*GP*TP*CP*AP*T)-3', Transcriptional regulator | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
1MU9
 
 | | Crystal Structure of a Human Tyrosyl-DNA Phosphodiesterase (Tdp1)-Vanadate Complex | | Descriptor: | GLYCEROL, Tyrosyl-DNA Phosphodiesterase, VANADATE ION | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 2002-09-23 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights Into Substrate Binding and Catalytic Mechanism of Human Tyrosyl-DNA Phosphodiesterase (Tdp1) from Vanadate- and Tungstate-Inhibited Structures
J.Mol.Biol., 324, 2002
|
|
6RVL
 
 | | Crystal structure of hCA II with Urea, N-(1,3-dihydro-1-hydroxy-2,1-benzoxaborol-6-yl)-?N'-phenyl- | | Descriptor: | 1-[1,1-bis(oxidanyl)-3~{H}-2,1-benzoxaborol-1-ium-6-yl]-3-phenyl-thiourea, Carbonic anhydrase 2, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exploring benzoxaborole derivatives as carbonic anhydrase inhibitors: a structural and computational analysis reveals their conformational variability as a tool to increase enzyme selectivity.
J Enzyme Inhib Med Chem, 34, 2019
|
|
5A13
 
 | |
3VOX
 
 | |
2BTN
 
 | | Crystal Structure and Catalytic Mechanism of the Quorum-Quenching N- Acyl Homoserine Lactone Hydrolase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, ZINC ION | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Lee, J.S, Kang, B.S, Kim, K.J, Derewenda, Z.S, Oh, T.K, Lee, C.H, Lee, J.K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5WTP
 
 | | Crystal structure of the C-terminal domain of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | OmpA family protein, SULFATE ION | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
3VPY
 
 | | Crystal structure of Arabidopsis DDL FHA domain | | Descriptor: | FHA domain-containing protein DDL | | Authors: | Yuan, Y.A, Machida, S. | | Deposit date: | 2012-03-15 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arabidopsis thaliana Dawdle Forkhead-Associated Domain reveals a conserved phospho-threonine recognition cleft for Dicer-like1 binding.
Mol Plant, 2013
|
|
3VQD
 
 | | HIV-1 IN core domain in complex with 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid | | Descriptor: | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
