4QYX
 
 | | Crystal structure of YDR533Cp | | Descriptor: | Probable chaperone protein HSP31 | | Authors: | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2A24
 
 | | HADDOCK Structure of HIF-2a/ARNT PAS-B Heterodimer | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain protein 1 | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-06-21 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization.
J.Mol.Biol., 353, 2005
|
|
2KRG
 
 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
5FNZ
 
 | |
6LOG
 
 | |
1QDV
 
 | | N-TERMINAL DOMAIN, VOLTAGE-GATED POTASSIUM CHANNEL KV1.2 RESIDUES 33-131 | | Descriptor: | KV1.2 VOLTAGE-GATED POTASSIUM CHANNEL | | Authors: | Minor Jr, D.L, Lin, Y.-F, Mobley, B.C, Yu, M, Jan, Y.N, Jan, L.Y, Berger, J.M. | | Deposit date: | 1999-07-10 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel.
Cell(Cambridge,Mass.), 102, 2000
|
|
1GQE
 
 | |
1QDW
 
 | | N-TERMINAL DOMAIN, VOLTAGE-GATED POTASSIUM CHANNEL KV1.2 RESIDUES 33-119 | | Descriptor: | KV1.2 VOLTAGE-GATED POTASSIUM CHANNEL | | Authors: | Minor Jr, D.L, Lin, Y.-F, Mobley, B.C, Avelar, A, Jan, Y.N, Jan, L.Y, Berger, J.M. | | Deposit date: | 1999-07-10 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel.
Cell(Cambridge,Mass.), 102, 2000
|
|
8CP5
 
 | |
8CP2
 
 | |
5XD9
 
 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
1HF0
 
 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | Descriptor: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
5BXX
 
 | | Crystal structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
6O47
 
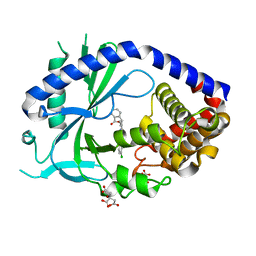 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | Descriptor: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EFE
 
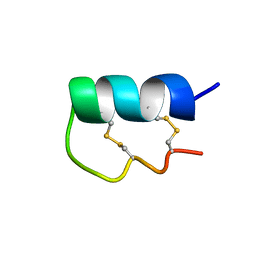 | | NMR Solution Structure of vil14a | | Descriptor: | Kappa-conotoxin vil14a | | Authors: | Dovell, S, Mari, F, Moller, C, Melaun, C. | | Deposit date: | 2018-08-16 | | Release date: | 2018-09-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Definition of the R-superfamily of conotoxins: Structural convergence of helix-loop-helix peptidic scaffolds.
Peptides, 107, 2018
|
|
5UNQ
 
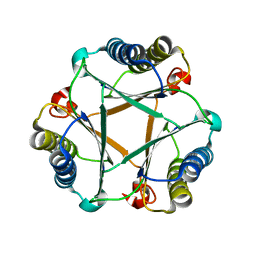 | |
2PJY
 
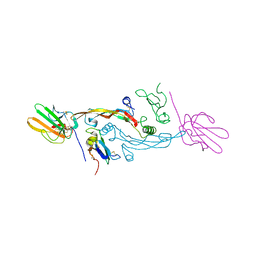 | |
6FI9
 
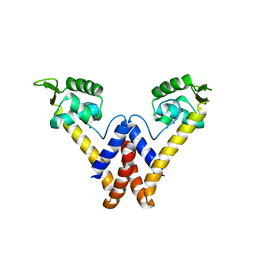 | |
7ALN
 
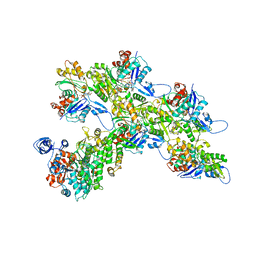 | | Cryo-EM structure of the divergent actomyosin complex from Plasmodium falciparum Myosin A in the Rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Robert-Paganin, J, Xu, X.-P, Swift, M.F, Auguin, D, Robblee, J.P, Lu, H, Fagnant, P.M, Krementsova, E.B, Trybus, K.M, Houdusse, A, Volkmann, N, Hanein, D. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity.
Nat Commun, 12, 2021
|
|
2KQM
 
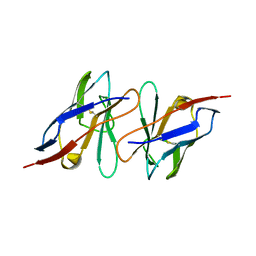 | |
2LKY
 
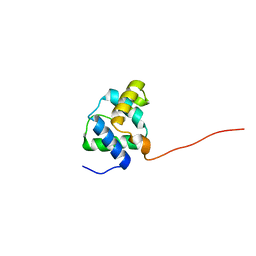 | |
2KQN
 
 | |
2M0N
 
 | |
6QYQ
 
 | | Crystal structure of human thymidylate synthase (hTS) variant R175C | | Descriptor: | CHLORIDE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Functional Characterization of the Human Thymidylate Synthase (hTS) Interface Variant R175C, New Perspectives for the Development of hTS Inhibitors.
Molecules, 24, 2019
|
|
6VIE
 
 | | Structure of caspase-1 in complex with gasdermin D | | Descriptor: | Caspase-1 subunit p10, Caspase-1 subunit p20, Gasdermin-D | | Authors: | Liu, Z, Xiao, T.S. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage.
Immunity, 53, 2020
|
|
