1B1B
 
 | | IRON DEPENDENT REGULATOR | | Descriptor: | PROTEIN (IRON DEPENDENT REGULATOR), SULFATE ION, ZINC ION | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1998-11-19 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied.
J.Mol.Biol., 285, 1999
|
|
6HL0
 
 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide | | Descriptor: | Bile acid receptor, NCoA-2 peptide (Nuclear receptor coactivator 2), LYS-GLU-ASN-ALA-LEU-LEU-ARG-TYR-LEU-LEU-ASP-LYS-ASP | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
6WP7
 
 | |
3NAU
 
 | | Crystal structure of ZHX2 HD2 (zinc-fingers and homeoboxes protein 2, homeodomain 2) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 2 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
7QFZ
 
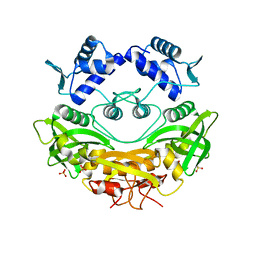 | | BrxR, a WYL-domain containing transcriptional regulator | | Descriptor: | SULFATE ION, WYL domain-containing protein | | Authors: | Picton, D.M, Blower, T.R. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A widespread family of WYL-domain transcriptional regulators co-localizes with diverse phage defence systems and islands.
Nucleic Acids Res., 50, 2022
|
|
7XI5
 
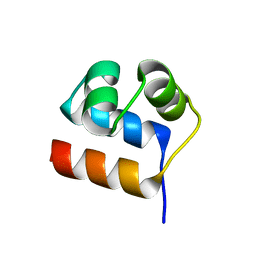 | | Anti-CRISPR-associated Aca10 | | Descriptor: | Transcriptional regulator | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2022-04-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of anti-CRISPR operon repression by Aca10.
Nucleic Acids Res., 50, 2022
|
|
7LS1
 
 | | 80S ribosome from mouse bound to eEF2 (Class II) | | Descriptor: | 28S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7LS2
 
 | | 80S ribosome from mouse bound to eEF2 (Class I) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
4BL8
 
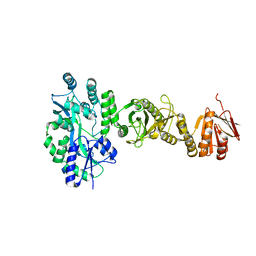 | | Crystal structure of full-length human Suppressor of fused (SUFU) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Karlstrom, M, Finta, C, Cherry, A.L, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6HL1
 
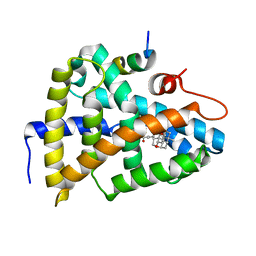 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide and CDCA | | Descriptor: | Bile acid receptor, CHENODEOXYCHOLIC ACID, NCoA-2 peptide (Nuclear receptor coactivator 2), ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
7N5T
 
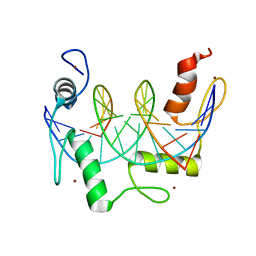 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 5) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7N5S
 
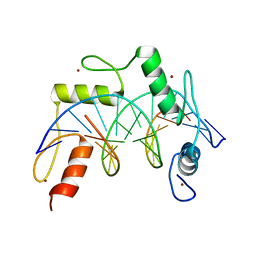 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 6) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7YTA
 
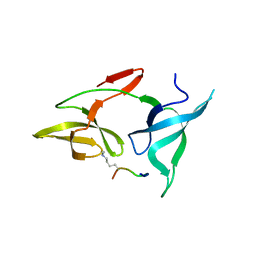 | |
7KW7
 
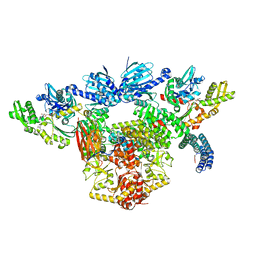 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
7SME
 
 | | p107 pocket domain complexed with HDAC1 peptide | | Descriptor: | Histone deacetylase 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7TDP
 
 | |
7SMD
 
 | | p107 pocket domain complexed with EID1 peptide | | Descriptor: | EP300-interacting inhibitor of differentiation 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SMF
 
 | | p107 pocket domain complexed with mutated HDAC1-3X peptide | | Descriptor: | Histone deacetylase 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SMC
 
 | | p107 pocket domain complexed with ARID4A peptide | | Descriptor: | AT-rich interactive domain-containing protein 4A, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
4UVA
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1R,2S) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS)-4a-[(1S,3E)-3-imino-1-phenylbutyl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
5MYT
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK921295A at 1.61A resolution | | Descriptor: | 4-methylsulfanyl-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5K5J
 
 | |
4A12
 
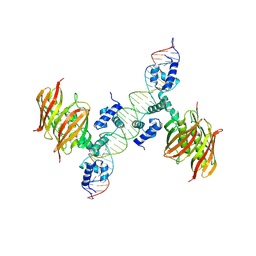 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with DNA operator | | Descriptor: | FAPR PROMOTER, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
7TDV
 
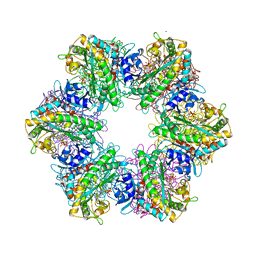 | |
7TEN
 
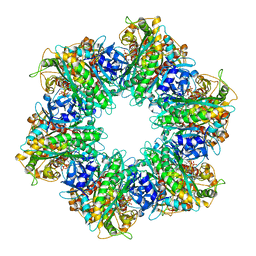 | |
