8FC8
 
 | | Cryo-EM structure of the human TRPV4 in complex with GSK1016790A | | Descriptor: | CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
6HHI
 
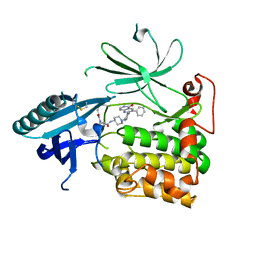 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 30b | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[1-[[4-(5-oxidanylidene-3-phenyl-6~{H}-1,6-naphthyridin-2-yl)phenyl]methyl]piperidin-4-yl]-3-(propanoylamino)benzamide | | Authors: | Landel, I, Weisner, J, Mueller, M.P, Scheinpflug, R, Rauh, D. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and chemical insights into the covalent-allosteric inhibition of the protein kinase Akt.
Chem Sci, 10, 2019
|
|
8FCB
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK1016790A | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
1Y1A
 
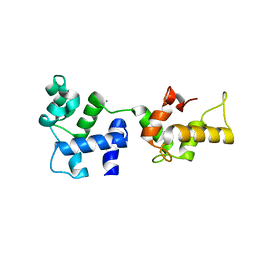 | | CRYSTAL STRUCTURE OF CALCIUM AND INTEGRIN BINDING PROTEIN | | Descriptor: | CALCIUM ION, Calcium and integrin binding 1 (calmyrin), GLUTATHIONE | | Authors: | Blamey, C.J, Ceccarelli, C, Naik, U.P, Bahnson, B.J. | | Deposit date: | 2004-11-17 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of calcium- and integrin-binding protein 1: Insights into redox regulated functions
Protein Sci., 14, 2005
|
|
6JP5
 
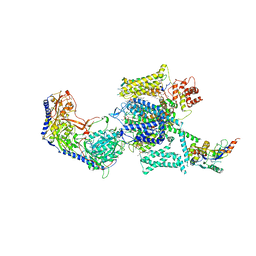 | | Rabbit Cav1.1-Nifedipine Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
1YHP
 
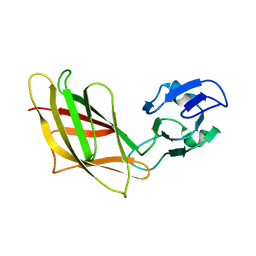 | | Solution Structure of Ca2+-free DdCAD-1 | | Descriptor: | Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YMG
 
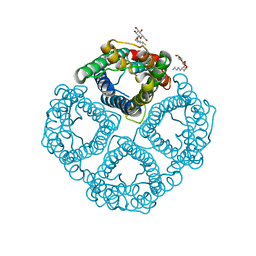 | | The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution | | Descriptor: | Lens fiber major intrinsic protein, nonyl beta-D-glucopyranoside | | Authors: | Harries, W.E.C, Akhavan, D, Miercke, L.J.W, Khademi, S, Stroud, R.M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6TV7
 
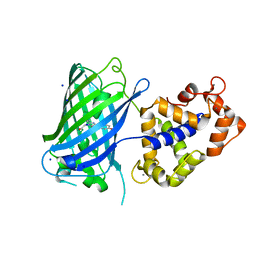 | | Crystal structure of rsGCaMP in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, SODIUM ION, rsGCaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-01-09 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
4IK3
 
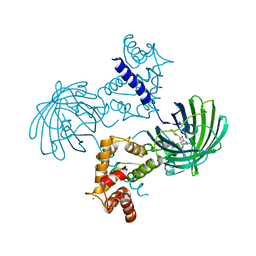 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
3BZ7
 
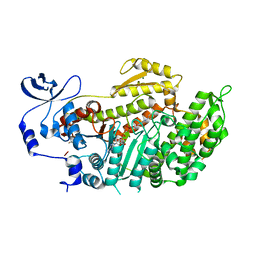 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | Descriptor: | (3aS)-3a-hydroxy-5-methyl-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Allingham, J.S, Rayment, I. | | Deposit date: | 2008-01-17 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
6VE7
 
 | | The inner junction complex of Chlamydomonas reinhardtii doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 20, FAP276, FAP52, ... | | Authors: | Khalifa, A.A.Z, Ichikawa, M, Bui, K.H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications.
Elife, 9, 2020
|
|
7AUG
 
 | | Crystal structure of rsGCamP1.3 in the ON state | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
3DFC
 
 | |
3B3N
 
 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
3BZ9
 
 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | Descriptor: | (3aS)-3a-hydroxy-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Rayment, I. | | Deposit date: | 2008-01-17 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
3D2F
 
 | | Crystal structure of a complex of Sse1p and Hsp70 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
3DGK
 
 | |
3BZ8
 
 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | Descriptor: | (3aS)-3a-hydroxy-7-methyl-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Rayment, I. | | Deposit date: | 2008-01-17 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
3CS1
 
 | | Flagellar Calcium-binding Protein (FCaBP) from T. cruzi | | Descriptor: | Flagellar calcium-binding protein | | Authors: | Ames, J.B, Ladner, J.E, Wingard, J.N, Robinson, H, Fisher, A. | | Deposit date: | 2008-04-08 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Membrane Targeting by the Flagellar Calcium-binding Protein (FCaBP), a Myristoylated and Palmitoylated Calcium Sensor in Trypanosoma cruzi.
J.Biol.Chem., 283, 2008
|
|
6WOU
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 mutant R176Q in complex with FKBP12.6 in nanodisc | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
3DQS
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
4JWQ
 
 | | Crystal Structure of the Calcium Binding Domain of CDPK3 from Plasmodium Berghei, PB000947.00 | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, SULFATE ION | | Authors: | Wernimont, A.K, Loppnau, P, Lin, Y.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Hui, R, Mottaghi, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-27 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Calcium Binding Domain of CDPK3 from Plasmodium Berghei, PB000947.00
TO BE PUBLISHED
|
|
6WOV
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 wild type in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Nayak, A.R, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
3B3P
 
 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3BAT
 
 | |
