7JHF
 
 | |
7JGY
 
 | |
8EZV
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EZZ
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a2 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-difluoroazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F2D
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML4006a | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopiperidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F02
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a4 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F2C
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML3006a | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopyrrolidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
5CUL
 
 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
5CUK
 
 | | Crystal structure of the PscP SS domain | | Descriptor: | GLYCEROL, Ruler protein | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
6TAM
 
 | |
5E96
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 6'-acetyltransferase, PHOSPHATE ION | | Authors: | Berghuis, A.M, Burk, D.L, Baettig, O.M, Shi, K. | | Deposit date: | 2015-10-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive characterization of ligand-induced plasticity changes in a dimeric enzyme.
Febs J., 283, 2016
|
|
7QBZ
 
 | | Crystal structure Cadmium translocating P-type ATPase | | Descriptor: | Cadmium translocating P-type ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7QC0
 
 | | Crystal structure of Cadmium translocating P-type ATPase | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cadmium translocating P-type ATPase, MAGNESIUM ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
6N8F
 
 | |
7R60
 
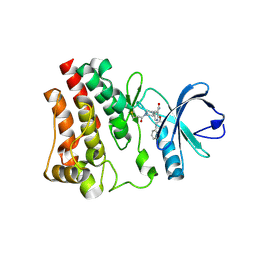 | | BTK in complex with 18A | | Descriptor: | 2-(4-phenoxyphenoxy)-5-[(3R)-1-(prop-2-enoyl)piperidin-3-yl]pyridine-3-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
7R61
 
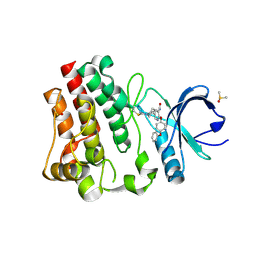 | | BTK in complex with 25A | | Descriptor: | 5-{(3S)-1-[(Z)-iminomethyl]piperidin-3-yl}-2-(4-phenoxyphenoxy)pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
6N8H
 
 | |
6N8I
 
 | |
5A89
 
 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with FMN and ADP(P 21 21 21) | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5ABA
 
 | |
5A88
 
 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
5AOI
 
 | |
5AB9
 
 | |
5A4J
 
 | | Crystal structure of FTHFS1 from T.acetoxydans Re1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, D(-)-TARTARIC ACID, ... | | Authors: | Bergdahl, R, Jacobson, F, Muller, B, Mikkelsen, N, Schurer, A, Sandgren, M. | | Deposit date: | 2015-06-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization, Crystallization and Three- Dimensional Structures of Formyltetrahydrofolate Synthetase (Fthfs) from the Syntrophic Acetate Oxidising Bacterium Tepidanaerobacter Acetatoxydans Re1
To be Published
|
|
