2EN5
 
 | |
7D3E
 
 | | Cryo-EM structure of human DUOX1-DUOXA1 in low-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
2EK2
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (E140M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Tanaka, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (E140M)
To be Published
|
|
7D78
 
 | | The structure of thioesterase DcsB | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96543121 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
5WPQ
 
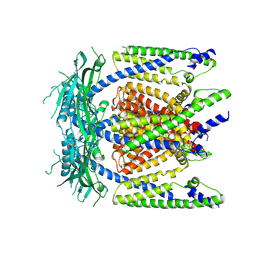 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed I conformation at 3.64 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
2E7Z
 
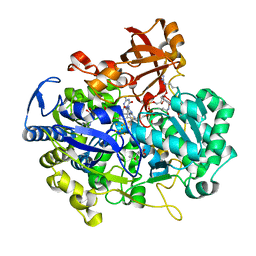 | | Acetylene Hydratase from Pelobacter acetylenicus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Einsle, O, Kroneck, P.M.H, Seiffert, G.B, Messerschmidt, A. | | Deposit date: | 2007-01-15 | | Release date: | 2007-02-27 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of the non-redox-active tungsten/[4Fe:4S] enzyme acetylene hydratase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7DJQ
 
 | |
5UDY
 
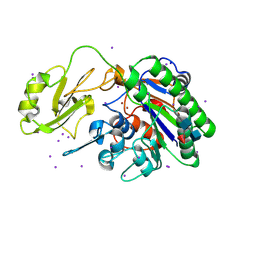 | | Human alkaline sphingomyelinase (alk-SMase, ENPP7, NPP7) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Liu, F, Illes, K, Nagar, B. | | Deposit date: | 2016-12-28 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human alkaline sphingomyelinase provides insights into substrate recognition.
J. Biol. Chem., 292, 2017
|
|
7CY3
 
 | |
7CY9
 
 | |
2EK7
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M) | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M)
To be Published
|
|
7D2A
 
 | | CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
4ZG9
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(11aS)-6-(4-fluorobenzyl)-1,3-dioxo-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indol-2(3H)-yl]propanoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
8DSC
 
 | | Human NAMPT in complex with substrate NAM and small molecule activator NP-A1-R | | Descriptor: | (3R)-1-[2-(4-methylphenyl)-2H-pyrazolo[3,4-d]pyrimidin-4-yl]-N-{[4-(methylsulfanyl)phenyl]methyl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
7CY1
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C, SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 2021
|
|
6SGD
 
 | | Nek2 kinase covalently bound to 2-arylamino-6-ethynylpurine inhibitor 24 | | Descriptor: | 4-[(6-ethenyl-7~{H}-purin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SODIUM ION, ... | | Authors: | Richards, M.W, Mas-Droux, C.P, Bayliss, R. | | Deposit date: | 2019-08-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Arylamino-6-ethynylpurines are cysteine-targeting irreversible inhibitors of Nek2 kinase.
Rsc Med Chem, 11, 2020
|
|
7D3F
 
 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7D79
 
 | | The structure of DcsB complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.10411429 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
6SAU
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
4WMU
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 2 AT 1.55A | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Faiman, J.W. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6STX
 
 | | Copper oxidase from Colletotrichum graminicola | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Kelch domain-containing protein, ... | | Authors: | Offen, W.A, Henrissat, B, Davies, G.J. | | Deposit date: | 2019-09-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Fungal Copper Radical Oxidase with High Catalytic Efficiency toward 5-Hydroxymethylfurfural and Benzyl Alcohols for Bioprocessing
Acs Catalysis, 2020
|
|
2EJZ
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (Y11M) | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Asada, Y, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (Y11M)
To be Published
|
|
2EH4
 
 | |
2EKB
 
 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M)
To be Published
|
|
7DTB
 
 | |
