7FS9
 
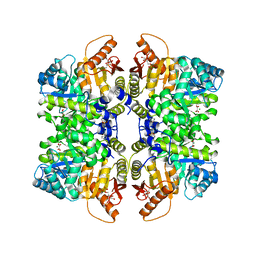 | | Structure of liver pyruvate kinase in complex with allosteric modulator 22 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(4-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSD
 
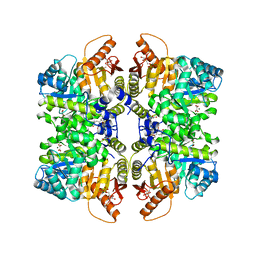 | | Structure of liver pyruvate kinase in complex with allosteric modulator 44 | | Descriptor: | (1M)-N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)-3',4,4',5-tetrahydroxy[1,1'-biphenyl]-2-sulfonamide, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS2
 
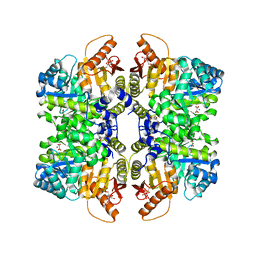 | | Structure of liver pyruvate kinase in complex with allosteric modulator 13 | | Descriptor: | (1P)-3',4,4',5-tetrahydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}[1,1'-biphenyl]-2-sulfonamide, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS8
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 21 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(3-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSC
 
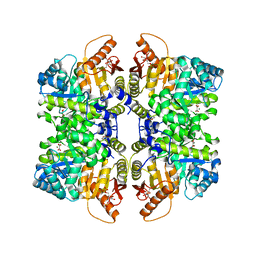 | | Structure of liver pyruvate kinase in complex with allosteric modulator 42 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)benzenesulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7MSJ
 
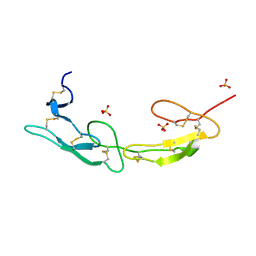 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
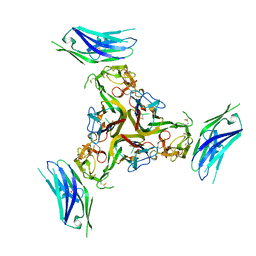 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
1QRY
 
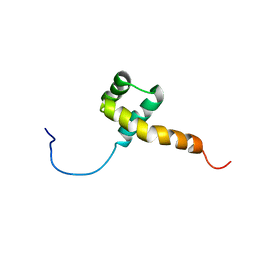 | | Homeobox protein VND (ventral nervous system defective protein) | | Descriptor: | PROTEIN (HOMEOBOX VENTRAL NERVOUS SYSTEM DEFECTIVE PROTEIN) | | Authors: | Xiang, B. | | Deposit date: | 1999-06-16 | | Release date: | 1999-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Distortion of the three-dimensional structure of the vnd/NK-2 homeodomain bound to DNA induced by an embryonically lethal A35T point mutation.
Biochemistry, 42, 2003
|
|
2KM0
 
 | | Cu(I)-bound CopK | | Descriptor: | COPPER (I) ION, Copper resistance protein K | | Authors: | Bersch, B. | | Deposit date: | 2009-07-15 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CopK from Cupriavidus metallidurans CH34 Binds Cu(I) in a Tetrathioether Site: Characterization by X-ray Absorption and NMR Spectroscopy
J.Am.Chem.Soc., 2010
|
|
2KN4
 
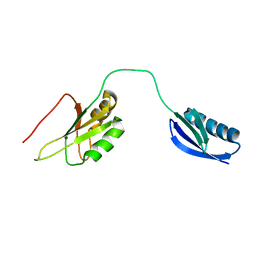 | | The structure of the RRM domain of SC35 | | Descriptor: | Immunoglobulin G-binding protein G,Serine/arginine-rich splicing factor 2 | | Authors: | Clayton, J.C, Goult, B.T, Lian, L.-Y. | | Deposit date: | 2009-08-14 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure and selectivity of the SR protein SRSF2 RRM domain with RNA
Nucleic Acids Res., 2011
|
|
2MXB
 
 | |
3H1J
 
 | | Stigmatellin-bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
4YVY
 
 | |
4YX9
 
 | |
4FK8
 
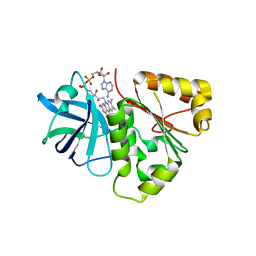 | |
6WXN
 
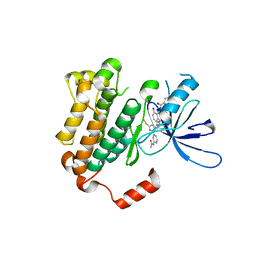 | | EGFR(T790M/V948R) in complex with LN3844 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
5VSG
 
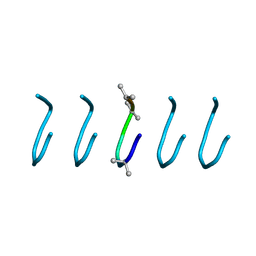 | | Fibrils of the super helical repeat peptide, SHR-FF, grown at elevated temperature | | Descriptor: | Super Helical Repeat Peptide SHR-FF | | Authors: | Mondal, S, Sawaya, M.R, Eisenberg, D.S, Gazit, E. | | Deposit date: | 2017-05-11 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Transition of Metastable Cross-alpha Crystals into Cross-beta Fibrils by beta-Turn Flipping.
J.Am.Chem.Soc., 141, 2019
|
|
4ZEV
 
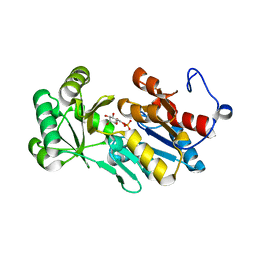 | | Crystal structure of PfHAD1 in complex with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZAK
 
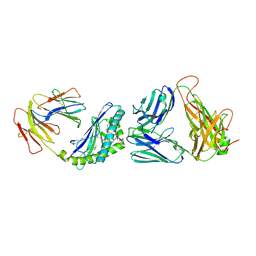 | | Crystal structure of the mCD1d/DB06-1/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Birkholz, A.M. | | Deposit date: | 2015-04-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production.
J Immunol., 195, 2015
|
|
2H1Z
 
 | |
2KSK
 
 | |
1VIV
 
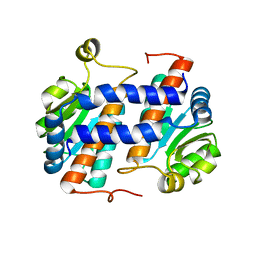 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical protein yckF | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VI2
 
 | | Crystal structure of shikimate-5-dehydrogenase with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, Shikimate 5-dehydrogenase 2 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIM
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | FORMIC ACID, Hypothetical protein AF1796 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VGZ
 
 | |
