8B7Y
 
 | | Cryo-EM structure of the E.coli 70S ribosome in complex with the antibiotic Myxovalargin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Koller, T.O, Graf, M, Wilson, D.N. | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
6TTC
 
 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
1HLF
 
 | | BINDING OF GLUCOPYRANOSYLIDENE-SPIRO-THIOHYDANTOIN TO GLYCOGEN PHOSPHORYLASE B: KINETIC AND CRYSTALLOGRAPHIC STUD | | Descriptor: | (5S,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2-thioxo-6-oxa-1,3-diazaspiro[4.5]decan-4-one, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Skamnaki, V.T, Docsa, T, Toth, B, Gergely, P, Osz, E, Szilagyi, L, Somsak, L. | | Deposit date: | 2000-12-01 | | Release date: | 2000-12-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranosylidene spirothiohydantoin
binding to glycogen phosphorylase B
BIOORG.MED.CHEM., 10, 2002
|
|
4ZIL
 
 | | Crystal structure of Rv2466c and oxidoreductase from Mycobacterium tuberculosis in its reduced state | | Descriptor: | DSBA oxidoreductase, MAGNESIUM ION | | Authors: | Albesa-Jove, D, Urresti, S, Comino, N, Tersa, M, Guerin, M.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | The Redox State Regulates the Conformation of Rv2466c to Activate the Antitubercular Prodrug TP053.
J.Biol.Chem., 290, 2015
|
|
6MDS
 
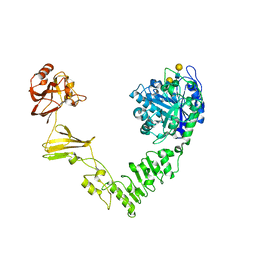 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with complex biantennary glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
8BDM
 
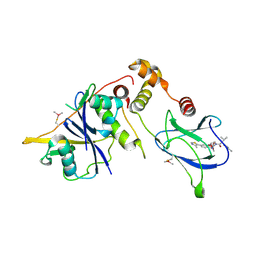 | | VCB in complex with compound 26 | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-(2-methoxyethoxy)-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8BDL
 
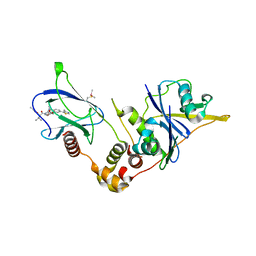 | | VCB in complex with compound 27 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-1-[2-(2-methoxyethoxy)-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8OW6
 
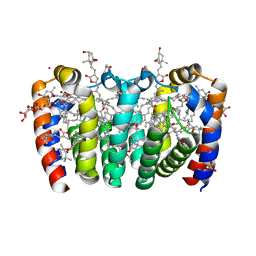 | |
8BDI
 
 | | VCB in complex with compound 32 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-3-(methylamino)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-3-oxidanylidene-propyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
5FU6
 
 | | NOT module of the human CCR4-NOT complex (Crystallization mutant) | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 2, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 3 | | Authors: | Raisch, T, Bhandari, D, Sabath, K, Helms, S, Valkov, E, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distinct Modes of Recruitment of the Ccr4-not Complex by Drosophila and Vertebrate Nanos
Embo J., 35, 2016
|
|
6CBQ
 
 | | Crystal structure of QscR bound to agonist S3 | | Descriptor: | (2S)-2-hexyl-N-[(3S)-2-oxooxolan-3-yl]decanamide, LuxR family transcriptional regulator | | Authors: | Churchill, M.E.A, Wysoczynski-Horita, C.L. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of agonism and antagonism of the Pseudomonas aeruginosa quorum sensing regulator QscR with non-native ligands.
Mol. Microbiol., 108, 2018
|
|
5VMY
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a hemi CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*CP*GP*CP*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
5H9F
 
 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
6E58
 
 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) | | Descriptor: | CALCIUM ION, Secreted Endo-beta-N-acetylglucosaminidase (EndoS) | | Authors: | Klontz, E.H, Trastoy, B, Gunther, S, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6MO7
 
 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6CSN
 
 | | Crystal structure of the designed light-gated anion channel iC++ at pH8.5 | | Descriptor: | CHLORIDE ION, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
1TM7
 
 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59Y mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-10 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
3NCG
 
 | |
5KND
 
 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
6MO8
 
 | | N-terminal bromodomain of human BRD2 in complex with 4,4'-(quinoline-5,7-diyl)bis(3,5-dimethylisoxazole) inhibitor | | Descriptor: | 5,7-bis(3,5-dimethyl-1,2-oxazol-4-yl)quinoline, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MR4
 
 | | Crystal structure of the Sth1 bromodomain from S.cerevisiae | | Descriptor: | Nuclear protein STH1/NPS1 | | Authors: | Seo, H.S, Hashimoto, H, Krolak, A, Debler, E.W, Blus, B.J. | | Deposit date: | 2018-10-11 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Substrate Affinity and Specificity of the ScSth1p Bromodomain Are Fine-Tuned for Versatile Histone Recognition.
Structure, 27, 2019
|
|
5VMW
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a double CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*(5CM)P*GP*(5CM)P*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
5C7D
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 17 | | Descriptor: | (2R)-4-[2-(6-chloro-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6DC7
 
 | | Apo Fab structure of mouse monoclonal antibody 8B2 | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Chukwu, J.E, Kong, X.-P. | | Deposit date: | 2018-05-04 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural characterization of monoclonal antibodies targeting C-terminal Ser404region of phosphorylated tau protein.
MAbs, 11, 2019
|
|
