1GD6
 
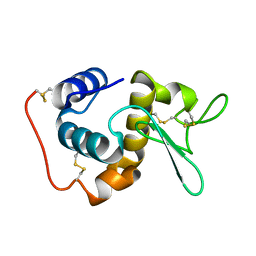 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | Deposit date: | 2000-09-19 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
2Q8I
 
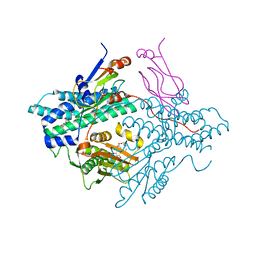 | | Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, GLYCEROL, ... | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
1FYC
 
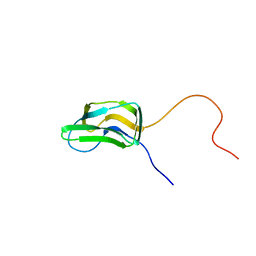 | | INNER LIPOYL DOMAIN FROM HUMAN PYRUVATE DEHYDROGENASE (PDH) COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE (E2P) | | Authors: | Howard, M.J, Fuller, C, Broadhurst, R.W, Quinn, J, Yeaman, S.J, Perham, R.N. | | Deposit date: | 1997-02-21 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the major autoantigen in primary biliary cirrhosis.
Gastroenterology, 115, 1998
|
|
5YGP
 
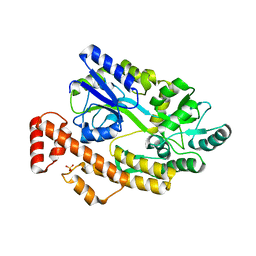 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5Y07
 
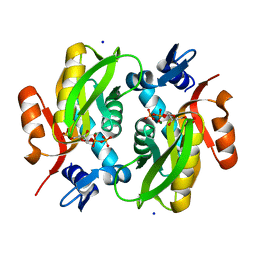 | |
5Y4A
 
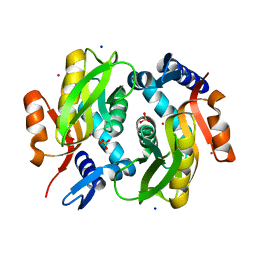 | |
5Y2G
 
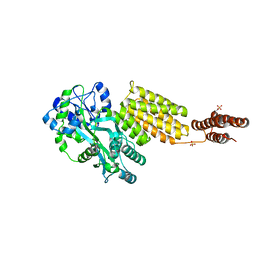 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7U0G
 
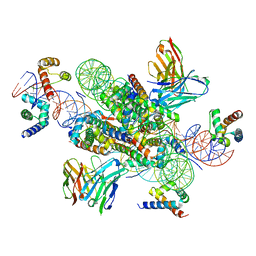 | | structure of LIN28b nucleosome bound 3 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
7U0I
 
 | | Structure of LIN28b nucleosome bound 2 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Tengfei, L, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
5YGS
 
 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6KEA
 
 | | crystal structure of MBP-tagged REV7-IpaB complex | | Descriptor: | Maltose-binding periplasmic protein,LINKER,hREV7,LINKER,Invasin IpaB,hREV3 | | Authors: | Wang, X, Pernicone, N, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
5ZOC
 
 | |
6JYZ
 
 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
5YW5
 
 | |
5ZCA
 
 | | Crystal structure of lambda repressor (1-20) fused with maltose-binding protein | | Descriptor: | CITRIC ACID, Repressor protein cI,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-02-16 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Co-translational folding of alpha-helical proteins: structural studies of intermediate-length variants of the lambda repressor
Febs Open Bio, 8, 2018
|
|
6K25
 
 | |
7UAJ
 
 | | Crystal structure of apo HPV16 E6 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein E6, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Shen, Q, Leonard, P.G, Cross, J.B. | | Deposit date: | 2022-03-13 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Disorder-to-order transition of the interdomain linker of HPV E6 upon E6AP binding reshapes p53 binding pocket
To Be Published
|
|
6KFQ
 
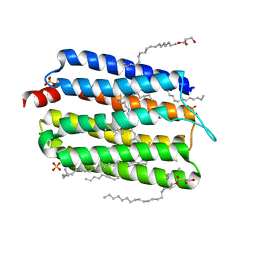 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
6K7E
 
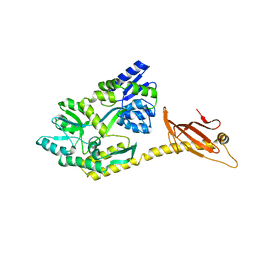 | |
5ZMI
 
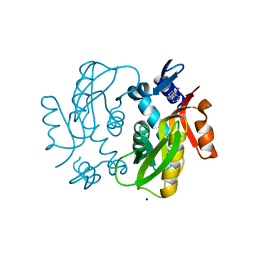 | |
5ZC7
 
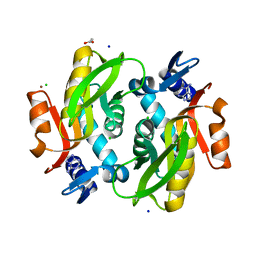 | |
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
6KXG
 
 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
7UN9
 
 | | SfSTING with c-di-GMP double fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
6LES
 
 | |
