4HIJ
 
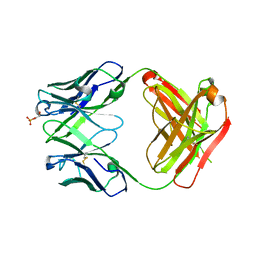 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound L-rhamnose-(1-2)-alpha-D-galactose-(3-O)-phosphate-2-glycerol | | Descriptor: | Fab 023.102 heavy chain, Fab 023.102 light chain, GLYCEROL, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
5KJ0
 
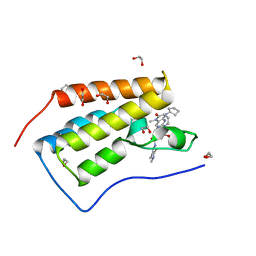 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH DB-1-264-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(7~{R})-8-cyclopentyl-7-ethyl-5-methyl-6-oxidanylidene-7~{H}-pteridin-2-yl]-methyl-amino]-3-methoxy-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W, Schonbrunn, E. | | Deposit date: | 2016-06-17 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assessment of Bromodomain Target Engagement by a Series of BI2536 Analogues with Miniaturized BET-BRET.
ChemMedChem, 11, 2016
|
|
6IXZ
 
 | |
4HIE
 
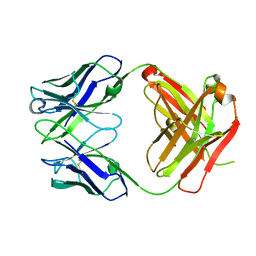 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 | | Descriptor: | Antibody 023.102, Fab 023.102 | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HII
 
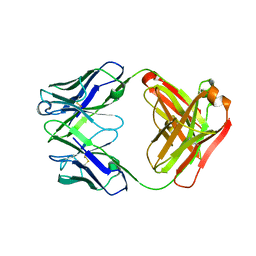 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose-galactose | | Descriptor: | Fab 023.102 heavy chain, Fab 023.102 light chain, alpha-L-rhamnopyranose-(1-2)-beta-D-galactopyranose | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
8CQH
 
 | |
6YLN
 
 | | mTurquoise2 SG P212121 - Directional optical properties of fluorescent proteins | | Descriptor: | EGFP, POTASSIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLU
 
 | | 14-3-3sigma in complex with BLNKpT152 phosphopeptide crystal structure | | Descriptor: | 14-3-3 protein sigma, BLNKpT152, MAGNESIUM ION | | Authors: | Soini, L, Leysen, S, Davis, J, Ottmann, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 14-3-3sigma in complex with BLNKpT152 phosphopeptide crystal structure
J.Struct.Biol., 2020
|
|
4HOT
 
 | | Crystal Structure of Full-Length Human IFIT5 with 5`-triphosphate Oligoadenine | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5, MAGNESIUM ION, RNA (5'-R(*(ATP)P*AP*AP*A)-3') | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
6JIK
 
 | | Aspergillus fumigatus Rho1 GsGTP | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Rho GTPase Rho1 | | Authors: | Bartual, S.G, Van Aalten, D.M.F. | | Deposit date: | 2019-02-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aspergillus fumigatus Rho1 gSGTP complex
To Be Published
|
|
5KVV
 
 | |
5KZ8
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 5,5-dimethyl-7-[(1~{S})-4-oxidanyl-1~{H}-inden-1-yl]-2-phenylazanyl-pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5KSX
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor AM-02-072 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [[(2~{S})-2-[[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoyl]amino]phosphonic acid | | Authors: | Park, J, Matralis, A, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
8D5G
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate | | Descriptor: | 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, CHLORIDE ION, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate
To Be Published
|
|
6YOY
 
 | |
6YP2
 
 | |
6YQT
 
 | | CRYSTAL STRUCTURE OF HUMAN CA II IN COMPLEX WITH A SULFONAMIDE DERIVATIVE OF 2-MERCAPTOBENZOXAZOLE | | Descriptor: | 2-sulfanylidene-3~{H}-1,3-benzoxazole-5-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2020-04-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 2-Mercaptobenzoxazoles: a class of carbonic anhydrase inhibitors with a novel binding mode to the enzyme active site.
Chem.Commun.(Camb.), 56, 2020
|
|
3ISC
 
 | | Binary complex of human DNA polymerase beta with an abasic site (THF) in the gapped DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(3DR)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
5KZA
 
 | | Crystal structure of the Rous sarcoma virus matrix protein (aa 2-102). Space group I41 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, virus matrix protein | | Authors: | Kingston, R.L, Chan, J, Vogt, V.M. | | Deposit date: | 2016-07-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cholesterol Promotes Protein Binding by Affecting Membrane Electrostatics and Solvation Properties.
Biophys. J., 113, 2017
|
|
4HIH
 
 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose. | | Descriptor: | Antibody 023.102, Fab 023.102, alpha-L-rhamnopyranose | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
6GZA
 
 | | Structure of murine leukemia virus capsid C-terminal domain | | Descriptor: | COBALT (II) ION, Capsid protein | | Authors: | Qu, K, Dolezal, M, Schur, F.K.M, Murciano, B, Rumlova, M, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6Y58
 
 | | Binary complex of 14-3-3 sigma (C38N) with the Estrogen Related Receptor gamma (LBD) phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Estrogen Related Receptor gamma phosphopeptide, MAGNESIUM ION | | Authors: | Somsen, B.A, Sijbesma, E, Leijten-van de Gevel, I.A, Ottmann, C. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
6YP3
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-028 | | Descriptor: | 1-(4-methylphenyl)-1,2,4-triazole, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Stabilizers of Protein-Protein Interactions through Imine-Based Tethering.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YP8
 
 | |
8DF2
 
 | |
