1V3D
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC2EN | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1V3U
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase in apo form | | Descriptor: | CHLORIDE ION, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
3HI9
 
 | |
1TLG
 
 | | STRUCTURE OF A TUNICATE C-TYPE LECTIN COMPLEXED WITH D-GALACTOSE | | Descriptor: | CALCIUM ION, POLYANDROCARPA LECTIN, ZINC ION, ... | | Authors: | Poget, S.F, Legge, G.B, Bycroft, M, Williams, R.L. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a tunicate C-type lectin from Polyandrocarpa misakiensis complexed with D -galactose.
J.Mol.Biol., 290, 1999
|
|
2QXL
 
 | | Crystal Structure Analysis of Sse1, a yeast Hsp110 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein homolog SSE1, MAGNESIUM ION, ... | | Authors: | Hendrickson, W.A, Liu, Q. | | Deposit date: | 2007-08-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insights into hsp70 chaperone activity from a crystal structure of the yeast hsp110 Sse1.
Cell(Cambridge,Mass.), 131, 2007
|
|
1X36
 
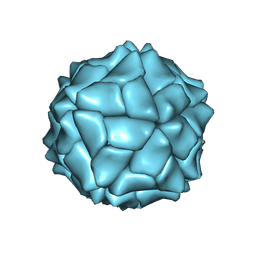 | | T=1 capsid of an amino-terminal deletion mutant of SeMV CP | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Sangita, V, Satheshkumar, P.S, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-04-29 | | Release date: | 2005-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a mutant T=1 capsid of Sesbania mosaic virus: role of water molecules in capsid architecture and integrity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3HKB
 
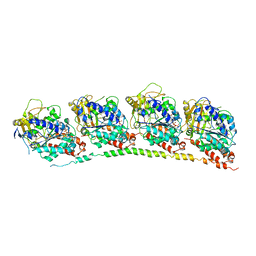 | | Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HKC
 
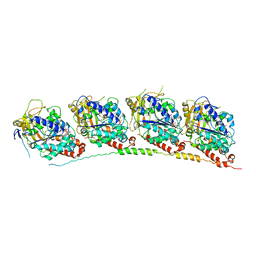 | | Tubulin-ABT751: RB3 stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HKE
 
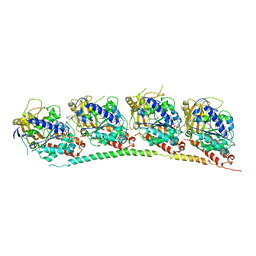 | | Tubulin-T138067: RB3 stathmin-like domain complex | | Descriptor: | 2,3,4,5,6-pentafluoro-N-(3-fluoro-4-methoxyphenyl)benzenesulfonamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HKD
 
 | | Tubulin-TN16 : RB3 stathmin-like domain complex | | Descriptor: | (3Z,5S)-5-benzyl-3-[1-(phenylamino)ethylidene]pyrrolidine-2,4-dione, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4BGP
 
 | | Crystal structure of La Crosse virus nucleoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, SULFATE ION | | Authors: | Reguera, J, Malet, H, Weber, F, Cusack, S. | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-24 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Encapsidation of Genomic RNA by La Crosse Orthobunyavirus Nucleoprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BHR
 
 | | Structure of the TTHA1221 type IV pilin protein from Thermus thermophilus | | Descriptor: | PILIN, TYPE IV, SULFATE ION | | Authors: | Karuppiah, V, Collins, R.F, Gao, Y, Thistlethwaite, A, Derrick, J.P. | | Deposit date: | 2013-04-05 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Assembly of an Inner Membrane Platform for Initiation of Type Iv Pilus Biogenesis
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BHQ
 
 | | Structure of the periplasmic domain of the PilN type IV pilus biogenesis protein from Thermus thermophilus | | Descriptor: | COMPETENCE PROTEIN PILN, MAGNESIUM ION | | Authors: | Karuppiah, V, Collins, R.F, Gao, Y, Thistlethwaite, A, Derrick, J.P. | | Deposit date: | 2013-04-05 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Assembly of an Inner Membrane Platform for Initiation of Type Iv Pilus Biogenesis
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MC2
 
 | |
1C0P
 
 | | D-AMINO ACIC OXIDASE IN COMPLEX WITH D-ALANINE AND A PARTIALLY OCCUPIED BIATOMIC SPECIES | | Descriptor: | D-ALANINE, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Umhau, S, Pollegioni, L, Molla, G, Diederichs, K, Welte, W, Pilone, S.M, Ghisla, S. | | Deposit date: | 1999-07-19 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C1L
 
 | | LACTOSE-LIGANDED CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I), beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1C4Z
 
 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
1BQM
 
 | | HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1C56
 
 | |
1BQN
 
 | | TYR 188 LEU HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1C4P
 
 | | BETA DOMAIN OF STREPTOKINASE | | Descriptor: | PROTEIN (STREPTOKINASE) | | Authors: | Wang, X, Zhang, X.C. | | Deposit date: | 1999-09-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of streptokinase beta-domain.
FEBS Lett., 459, 1999
|
|
2M8M
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
1C55
 
 | |
1CCM
 
 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin.
Proteins, 15, 1993
|
|
