2RSC
 
 | |
8GYA
 
 | |
2RVJ
 
 | |
2RN8
 
 | |
6K5M
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa (Rice) | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-05-29 | | Release date: | 2020-06-03 | | Last modified: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8H8I
 
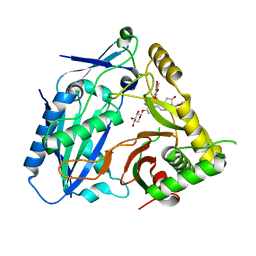 | | Triterpenoid saponin acetyltransferase, AmAT7-3 | | Descriptor: | AmAT7-3, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
6KHE
 
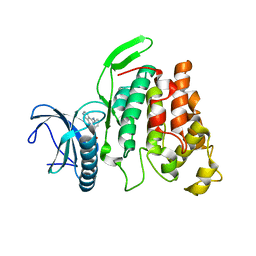 | | Crystal structure of CLK2 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
2RUG
 
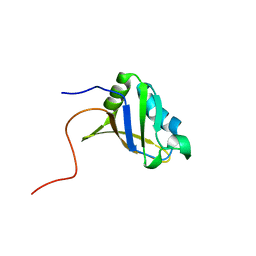 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
8GYE
 
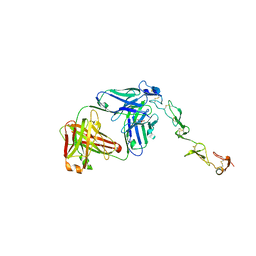 | |
2RLY
 
 | | FBP28WW2 domain in complex with PTPPPLPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14a
To Be Published
|
|
6KI6
 
 | | Crystal structure of BCL11A in complex with gamma-globin -115 HPFH region | | Descriptor: | B-cell lymphoma/leukemia 11A, DNA (5'-D(*AP*TP*AP*TP*TP*GP*GP*TP*CP*AP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*GP*AP*CP*CP*AP*AP*TP*A)-3'), ... | | Authors: | Li, F.D, Yang, Y, Shi, Y.Y. | | Deposit date: | 2019-07-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the recognition of gamma-globin gene promoter by BCL11A.
Cell Res., 29, 2019
|
|
2RSW
 
 | |
2RUR
 
 | | Solution structure of Human Pin1 PPIase C113S mutant | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
8GY8
 
 | |
2RVL
 
 | |
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RUN
 
 | | Solution Structure of Pre Transmembrane domain | | Descriptor: | Pre-transmembrane domain of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
1IX7
 
 | | Aspartate Aminotransferase Active Site Mutant V39F maleate complex | | Descriptor: | Aspartate Aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
2RSU
 
 | | Alternative structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Yagi-Utsumi, M, Kato, K, Kitahara, R. | | Deposit date: | 2012-06-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Q41N Variant of Ubiquitin as a Model for the Alternatively Folded N2 State of Ubiquitin
Biochemistry, 52, 2013
|
|
1IXR
 
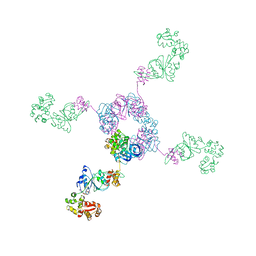 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
2RSX
 
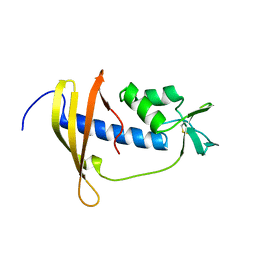 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
8GXH
 
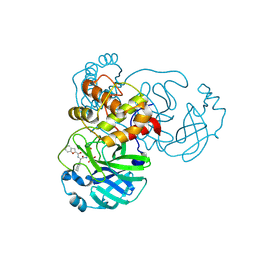 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14b
To Be Published
|
|
2RSE
 
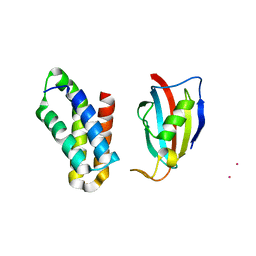 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
