8F0L
 
 | |
8ELA
 
 | | CTX-M-14 beta-lactamase mutant - N132A w MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lu, S, Palzkill, T, Hu, L, Prasad, B.V.V. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
6S21
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
8CLM
 
 | |
6XR1
 
 | |
1ADF
 
 | | CRYSTALLOGRAPHIC STUDIES OF TWO ALCOHOL DEHYDROGENASE-BOUND ANALOGS OF THIAZOLE-4-CARBOXAMIDE ADENINE DINUCLEOTIDE (TAD), THE ACTIVE ANABOLITE OF THE ANTITUMOR AGENT TIAZOFURIN | | Descriptor: | ALCOHOL DEHYDROGENASE, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ZINC ION | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Marquez, V.E, Carrell, H.L, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry, 33, 1994
|
|
1ANK
 
 | | THE CLOSED CONFORMATION OF A HIGHLY FLEXIBLE PROTEIN: THE STRUCTURE OF E. COLI ADENYLATE KINASE WITH BOUND AMP AND AMPPNP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Berry, M.B, Meador, B, Bilderback, T, Liang, P, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP.
Proteins, 19, 1994
|
|
7S1B
 
 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
8S9G
 
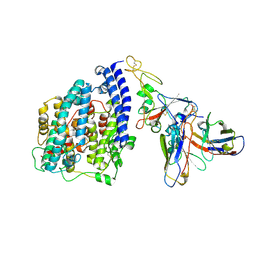 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6AH0
 
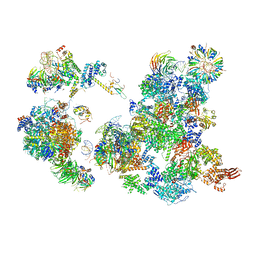 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6U0N
 
 | |
4WIF
 
 | |
6U6X
 
 | | Human SAMHD1 bound to deoxyribo(C*G*C*C*T)-oligonucleotide | | Descriptor: | DNA SC-GS-SC-SC-DT, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
6OI8
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with 7-((1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl)-6-(2-chloro-6-(pyridin-3-yl)phenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl]-6-[2-chloro-6-(pyridin-3-yl)phenyl]pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
6RJ2
 
 | | Crystal structure of PHGDH in complex with compound 40 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-[(1~{R})-1-[4-(ethanoylsulfamoyl)phenyl]ethyl]-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
4X4X
 
 | |
7S1C
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001897 (compound 1) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-3-ylphenyl)methanamine | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Sharma, P. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
103D
 
 | |
8OO1
 
 | |
6OJH
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
4WLV
 
 | | Crystal structure of NAD bound MDH2 | | Descriptor: | Malate dehydrogenase, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Eo, Y.M, Han, B.G, Ahn, H.C. | | Deposit date: | 2014-10-08 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NAD bound MDH2
To Be Published
|
|
7SR2
 
 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
1BC7
 
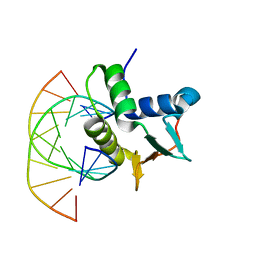 | | SERUM RESPONSE FACTOR ACCESSORY PROTEIN 1A (SAP-1)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*AP*GP*GP*AP*TP*GP*TP*G)-3'), PROTEIN (ETS-DOMAIN PROTEIN) | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1999-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
7SR1
 
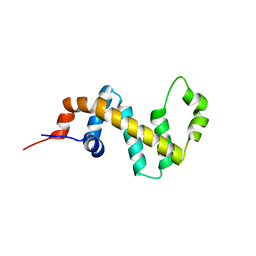 | |
6OI1
 
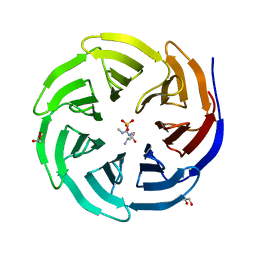 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
