8YB5
 
 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 187, 2024
|
|
8YB6
 
 | | Type I-EHNH Cascade complex | | Descriptor: | 61-nt crRNA, CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
8VZ8
 
 | | Crystal structure of mouse MAIT M2B TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
8VZ9
 
 | | Crystal structure of mouse MAIT M2A TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
8VZM
 
 | | DNA Ligase 1 captured with pre-step 3 ligation at the rA:T nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VZL
 
 | | DNA Ligase 1 captured with pre-step 3 ligation at the rG:C nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8YAX
 
 | | SARS-CoV-2 DMV nsp3-4 pore complex (full-pore) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-10 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8RYT
 
 | | Structural characterization of Thogoto Virus nucleoprotein provides insights into RNA encapsidation and assembly | | Descriptor: | Nucleoprotein | | Authors: | Roske, Y, Mikirtumov, V, Daumke, O, Kudryashev, M, Dick, A. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural characterization of Thogoto Virus nucleoprotein provides insights into viral RNA encapsidation and RNP assembly.
Structure, 32, 2024
|
|
8RYN
 
 | |
8RYQ
 
 | |
8RYM
 
 | |
8RYO
 
 | |
8RYP
 
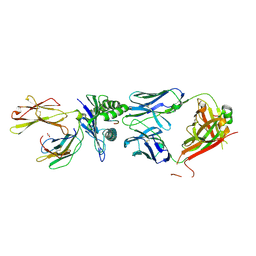 | |
8YAN
 
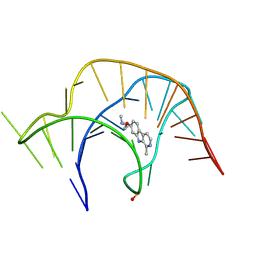 | |
8YAM
 
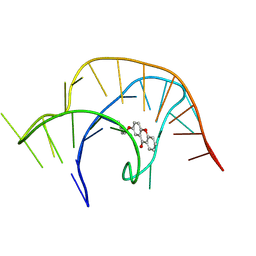 | |
8YAL
 
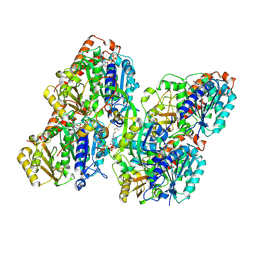 | | ATAT-2 bound K40Q MEC-12/MEC-7 microtubule | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
8RYS
 
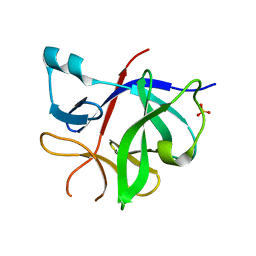 | | Human IL-1beta, unliganded | | Descriptor: | Interleukin-1 beta, SULFATE ION | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
8RYK
 
 | |
8YAJ
 
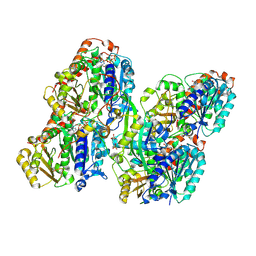 | | ATAT-2 bound MEC-12/MEC-7 microtubule without acetyl-CoA | | Descriptor: | Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
8YAR
 
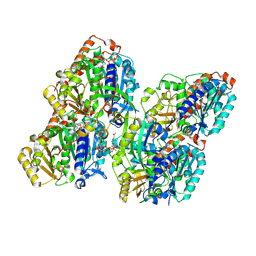 | | ATAT-2 bound K40R MEC-12/MEC-7 microtubule | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
8RXX
 
 | |
8VYL
 
 | | The structure of Human Hemoglobin in Complex with Nanobody BtNbE11 | | Descriptor: | ACETYL GROUP, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Grinter, R, Binks, S, Fox, D. | | Deposit date: | 2024-02-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The structure of a haemoglobin-nanobody complex reveals human beta-subunit-specific interactions.
Febs Lett., 598, 2024
|
|
8VYD
 
 | |
8YAA
 
 | | Cryo-EM structure of MIK2-SCOOP12-BAK1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1, ... | | Authors: | Jia, F.S, Xiao, Y, Chai, J.J. | | Deposit date: | 2024-02-08 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of MIK2-SCOOP12-BAK1
To Be Published
|
|
