4HG5
 
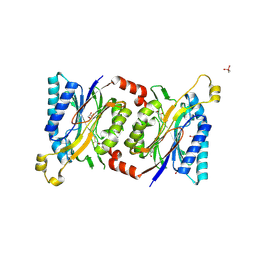 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with oxaloacetate | | Descriptor: | CACODYLATE ION, GLYCEROL, OXALOACETATE ION, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2BQX
 
 | |
8THC
 
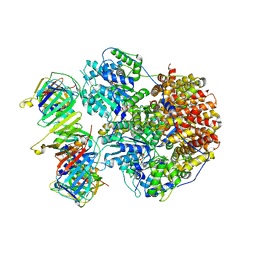 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
2C5O
 
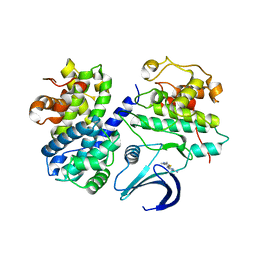 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
7QHG
 
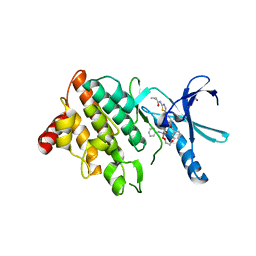 | | LIM domain kinase 2 (LIMK2) in complex with TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Salah, E, Hanke, T, Knapp, S. | | Deposit date: | 2021-12-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Development and Characterization of Type I, Type II, and Type III LIM-Kinase Chemical Probes.
J.Med.Chem., 65, 2022
|
|
2BSW
 
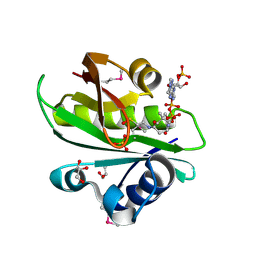 | | Crystal structure of a glyphosate-N-acetyltransferase obtained by DNA shuffling. | | Descriptor: | GLYCEROL, GLYPHOSATE N-ACETYLTRANSFERASE, OXIDIZED COENZYME A, ... | | Authors: | Keenan, R.J, Siehl, D.L, Gorton, R, Castle, L.A. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-08 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Shuffling as a Tool for Protein Crystallization.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BWA
 
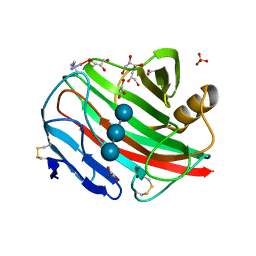 | |
2BKK
 
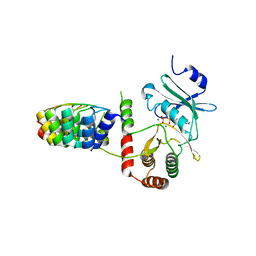 | | Crystal structure of Aminoglycoside Phosphotransferase APH(3')-IIIa in complex with the inhibitor AR_3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, DESIGNED ANKYRIN REPEAT INHIBITOR AR_3A, ... | | Authors: | Kohl, A, Amstutz, P, Parizek, P, Binz, H.K, Briand, C, Capitani, G, Forrer, P, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allosteric Inhibition of Aminoglycoside Phosphotransferase by a Designed Ankyrin Repeat Protein
Structure, 13, 2005
|
|
2BFE
 
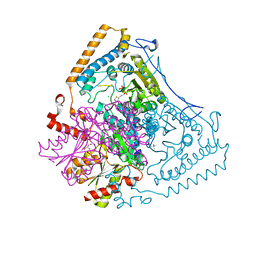 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFD
 
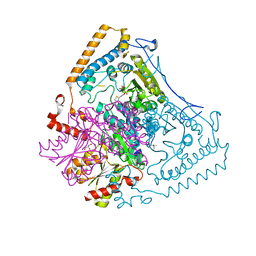 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2C2O
 
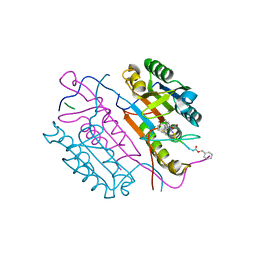 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-{4-[BENZYL(METHYL) AMINO]-4-OXOBUTANOYL}-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2C1E
 
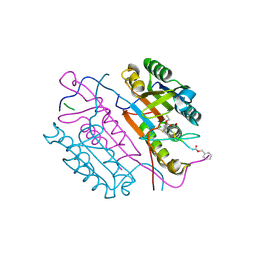 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Grutter, M.G. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2C2M
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-[4-(BENZYLOXY)-4-OXOBUTANOYL]-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14 -PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2C2K
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A.J, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
7L0E
 
 | |
2BFP
 
 | | Leishmania major pteridine reductase 1 in complex with NADP and tetrahydrobiopterin | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-10 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
7KSA
 
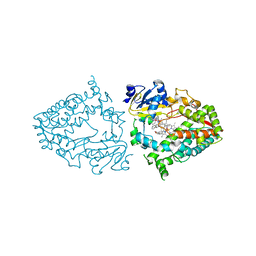 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (tert-butyl {1-[(1-oxo-3-phenyl-1-{[3-(pyridin-3-yl-kappaN)prop-1-en-1-yl]amino}propan-2-yl)sulfanyl]-3-phenylpropan-2-yl}carbamate)(6,6'-dimethyl-2,2'-bipyridine-kappa~2~N~1~,N~1'~)(1~2~,2~2~:2~6~,3~2~-terpyridine-kappa~3~N~1^{1~},N~2^{1~},N~3^{1~})ruthenium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.S. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photosensitive Ru(II) Complexes as Inhibitors of the Major Human Drug Metabolizing Enzyme CYP3A4.
J.Am.Chem.Soc., 143, 2021
|
|
7KS8
 
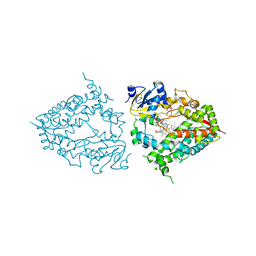 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (tert-butyl {1-[(3-oxo-3-{[(pyridin-3-yl-kappaN)methyl]amino}propyl)sulfanyl]-3-phenylpropan-2-yl}carbamate)(6,6'-dimethyl-2,2'-bipyridine-kappa~2~N~1~,N~1'~)(1~2~,2~2~:2~6~,3~2~-terpyridine-kappa~3~N~1^{1~},N~2^{1~},N~3^{1~})ruthenium, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I.S. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photosensitive Ru(II) Complexes as Inhibitors of the Major Human Drug Metabolizing Enzyme CYP3A4.
J.Am.Chem.Soc., 143, 2021
|
|
2BM0
 
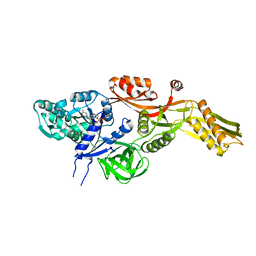 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant T84A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
2BM1
 
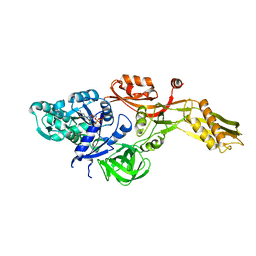 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant G16V | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
2BWY
 
 | | Glu383Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BOH
 
 | | Crystal structure of factor Xa in complex with compound "1" | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR XA | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-11 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BFA
 
 | | Leishmania major pteridine reductase 1 in complex with NADP and CB3717 | | Descriptor: | 1,2-ETHANEDIOL, 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
7KXW
 
 | | Crystal structure of DCLK1-KD in complex with DCLK1-IN-1 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-11-methyl-5-(2,2,2-trifluoroethyl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase DCLK1, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
2BWK
 
 | | Murine angiogenin, sulphate complex | | Descriptor: | ANGIOGENIN, SULFATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
