5LG3
 
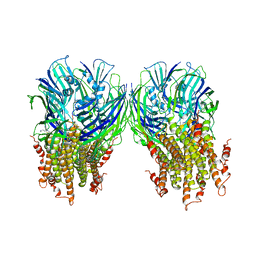 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.567 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1F1O
 
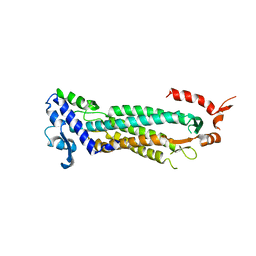 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
8QR0
 
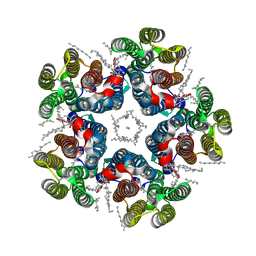 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
7ZJL
 
 | |
7PZD
 
 | | Cryo-EM structure of the NLRP3 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Hochheiser, I.V, Hagelueken, G, Behrmann, H, Behrmann, E, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation.
Sci Adv, 8, 2022
|
|
2WWA
 
 | | Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome | | Descriptor: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
6OWO
 
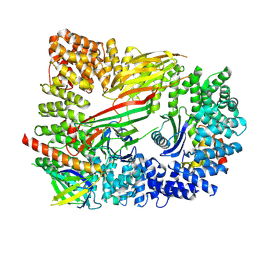 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 CORE BOUND TO NECAP | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J.S, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-09-11 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
6MCY
 
 | | Crystal structure of mouse Bak | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, FORMIC ACID | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis.
Nat.Chem.Biol., 15, 2019
|
|
8A62
 
 | | Small molecule stabilizer (compound 2) for FOXO1 and 14-3-3 | | Descriptor: | (3~{S})-1-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A6H
 
 | | Small molecule stabilizer (compound 7) for C-RAF and 14-3-3 | | Descriptor: | 14-3-3 protein sigma, 3-[2-(dimethylamino)ethyldisulfanyl]-1-[4-oxidanyl-4-[3-(trifluoromethyl)phenyl]piperidin-1-yl]propan-1-one, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A65
 
 | | Small molecule stabilizer (compound 3) for FOXO1 and 14-3-3 | | Descriptor: | (3~{S})-1-[2-azanyl-3,5-bis(chloranyl)phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A68
 
 | | Small molecule stabilizer (compound 5) for C-RAF(pS259) and 14-3-3 | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, RAF proto-oncogene serine/threonine-protein kinase, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
7Q69
 
 | | Crystal structure of Chaetomium thermophilum C30S Ahp1 in the pre-reaction state | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin domain-containing protein | | Authors: | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
8A6F
 
 | | Small molecule stabilizer (compound 8) for C-RAF and 14-3-3 | | Descriptor: | 1-[4-[4-chloranyl-3-(trifluoromethyl)phenyl]-4-oxidanyl-piperidin-1-yl]-3-[2-(dimethylamino)ethyldisulfanyl]propan-1-one, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6OXT
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-84 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY0
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-21 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6UYK
 
 | | Dark-operative protochlorophyllide oxidoreductase in the nucleotide-free form. | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein | | Authors: | Bacik, J.P, Imran, S.M.S, Watkins, M.B, Corless, E, Antony, E, Ando, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] cluster and released upon ATP binding.
J.Biol.Chem., 296, 2020
|
|
6OE8
 
 | | The crystal structure of hyper-thermostable AgUricase mutant K12C/E286C | | Descriptor: | MALONATE ION, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Shi, Y, Wang, T, Zhou, X.E, Liu, Q, Jiang, Y, Xu, H.E. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-based design of a hyperthermostable AgUricase for hyperuricemia and gout therapy.
Acta Pharmacol.Sin., 40, 2019
|
|
7T3Q
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active B state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
6ONC
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase produced without CooC, as-isolated | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE (III) ION, ... | | Authors: | Wittenborn, E.C, Cohen, S.E, Drennan, C.L. | | Deposit date: | 2019-04-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
7T4Q
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
6OF9
 
 | |
7T3T
 
 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3U
 
 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3R
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active C state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
