6NCP
 
 | | Crystal structure of HIV-1 broadly neutralizing antibody ACS202 | | Descriptor: | ACS202 Fab heavy chain, ACS202 Fab light chain, GLYCINE, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2018-12-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NK0
 
 | | EphA2 LBD in complex with bA-WLA-Yam peptide | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINOHEXANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lechtenberg, B.C, Pasquale, E.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Engineering nanomolar peptide ligands that differentially modulate EphA2 receptor signaling.
J.Biol.Chem., 294, 2019
|
|
9L7E
 
 | | Crystal structure of human kinesin-1 motor domain (G234A mutant) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-1 heavy chain, MAGNESIUM ION | | Authors: | Makino, T, Miyazono, K, Tanokura, M, Tomishige, M. | | Deposit date: | 2024-12-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tension-induced suppression of allosteric conformational changes coordinates kinesin-1 stepping.
J.Cell Biol., 224, 2025
|
|
4MYW
 
 | | Structure of HSV-2 gD bound to nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
4Y8X
 
 | |
8QLL
 
 | |
1THB
 
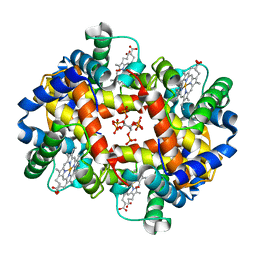 | |
7O1B
 
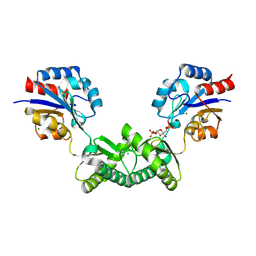 | | Human phosphomannomutase 2 (PMM2) wild-type co-crystallized with the activator glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ramon-Maiques, S, Briso-Montiano, A, Del Cano-Ochoa, F, Vilas, A, Perez, B, Rubio, V. | | Deposit date: | 2021-03-29 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures.
J Inherit Metab Dis, 45, 2022
|
|
4EM6
 
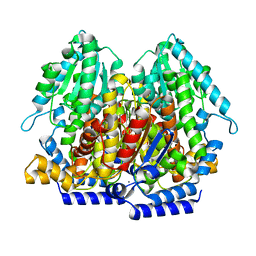 | |
6DCW
 
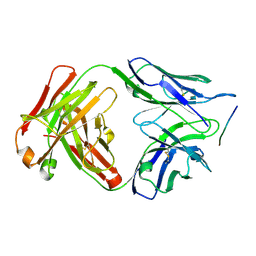 | |
6DD5
 
 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
9D4W
 
 | | Structure of PAK1 in complex with compound 12 | | Descriptor: | N~2~-{[(1s,4s)-4-aminocyclohexyl]methyl}-N~4~-[5-(trifluoromethyl)-1,3-thiazol-2-yl]pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Dementiev, A, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 127, 2025
|
|
9D4Y
 
 | | Structure of PAK1 in complex with compound 31 | | Descriptor: | N~2~-{[(1R,3R,4S)-4-amino-3-(3-chlorophenyl)cyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Fontano, E, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 127, 2025
|
|
9D4X
 
 | | Structure of PAK1 in complex with compound 16 | | Descriptor: | N~2~-{[(1R,3R,4S)-4-amino-3-(3-chlorophenyl)cyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Dementiev, A, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 127, 2025
|
|
4YPO
 
 | |
8QGH
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]-3-[(azanylidene-$l^{4}-azanylidene)amino]propanamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
9KUH
 
 | |
1TZD
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC CORE OF INOSITOL 1,4,5-TRISPHOSPHATE 3-KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MAGNESIUM ION | | Authors: | Miller, G.J, Hurley, J.H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase
Mol.Cell, 15, 2004
|
|
3TX7
 
 | | Crystal structure of LRH-1/beta-catenin complex | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Catenin beta-1, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Yumoto, F, Fletterick, R. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis of coactivation of liver receptor homolog-1 by beta-catenin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6NTT
 
 | |
2YNV
 
 | | Cys221 oxidized, Mono zinc GIM-1 - GIM-1-Ox. Crystal structures of Pseudomonas aeruginosa GIM-1: active site plasticity in metallo-beta- lactamases | | Descriptor: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
6NQG
 
 | |
7WX2
 
 | | CBP-BrD complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, CREB-binding protein | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
4Y8Y
 
 | |
4YWG
 
 | | Crystal structure of 830A in complex with V1V2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of anti-HIV-1 gp120 V1V2 antibody 830A, Light chain of anti-HIV-1 gp120 V1V2 antibody 830A, ... | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel.
J.Virol., 89, 2015
|
|
