3BQ3
 
 | | Crystal structure of S. cerevisiae Dcn1 | | Descriptor: | Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Chou, Y.C, Sicheri, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dcn1 Functions as a Scaffold-Type E3 Ligase for Cullin Neddylation.
Mol.Cell, 29, 2008
|
|
3WCG
 
 | | The complex structure of TcSQS with ligand, BPH1344 | | Descriptor: | Farnesyltransferase, putative, hydrogen [(1R)-2-(3-pentadecyl-1H-imidazol-3-ium-1-yl)-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | Descriptor: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
2N14
 
 | | Mdmx-295 | | Descriptor: | 4-({(4S,5R)-4-(3-chlorophenyl)-5-(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N0U
 
 | | Mdmx-057 | | Descriptor: | 4-[(4S,5R)-4-(3-chlorophenyl)-5-(4-chlorophenyl)-1-(3-oxidanylidenepiperazin-1-yl)carbonyl-4,5-dihydroimidazol-2-yl]-3-propan-2-yloxy-benzenecarbonitrile, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2MVV
 
 | | Solution Structure of the 5-phenyl-3-oxo-pentyl Actinorhodin Acyl Carrier Protein from Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, N~3~-{(2S)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-{2-[(3-oxo-5-phenylpentyl)sulfanyl]ethyl}-beta-alaninamide | | Authors: | Dong, X, Bailey, C, Williams, C, Crosby, J, Simpson, T.J, Willis, C.L, Crump, M.P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | ACP-ligand recognition: Selection of derivatized aromatic biosynthetic intermediates
To be Published
|
|
3WCE
 
 | | The complex structure of TcSQS with ligand, ER119884 | | Descriptor: | (3R)-3-{[2-benzyl-6-(3-methoxypropoxy)pyridin-3-yl]ethynyl}-1-azabicyclo[2.2.2]octan-3-ol, Farnesyltransferase, putative | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCM
 
 | | The complex structure of HsSQS wtih ligand, ER119884 | | Descriptor: | (3R)-3-{[2-benzyl-6-(3-methoxypropoxy)pyridin-3-yl]ethynyl}-1-azabicyclo[2.2.2]octan-3-ol, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-28 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCH
 
 | | The complex structure of HsSQS wtih ligand BPH1237 | | Descriptor: | Squalene synthase, hydrogen [(1R)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-hydroxy-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCJ
 
 | | The complex structure of HsSQS wtih ligand,E5700 | | Descriptor: | (3R)-3-({2-benzyl-6-[(3R,4S)-3-hydroxy-4-methoxypyrrolidin-1-yl]pyridin-3-yl}ethynyl)-1-azabicyclo[2.2.2]octan-3-ol, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCF
 
 | | The complex structure of HsSQS wtih ligand,BPH1218 | | Descriptor: | Squalene synthase, hydrogen [(1S)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
2MWY
 
 | | Mdmx-p53 | | Descriptor: | Cellular tumor antigen p53, Protein Mdm4 | | Authors: | Grace, C.R. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2VEE
 
 | | Structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2VEB
 
 | | High resolution structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | GLYCEROL, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2A7L
 
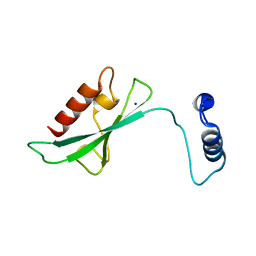 | | Structure of the human hypothetical ubiquitin-conjugating enzyme, LOC55284 | | Descriptor: | Hypothetical ubiquitin-conjugating enzyme LOC55284, SODIUM ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-05 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3HFU
 
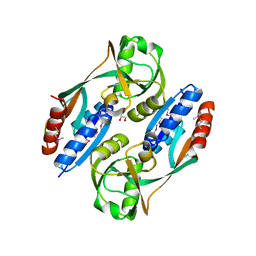 | | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, HTH-type transcriptional regulator cynR | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Dong, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide
TO BE PUBLISHED
|
|
3O3J
 
 | |
6IEN
 
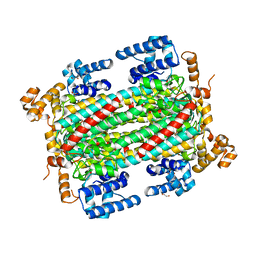 | | Substrate/product bound Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ARGININOSUCCINATE, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
2OM5
 
 | |
7BL8
 
 | | BAZ2A bromodomain in complex with the chemical probe BAZ2-ICR | | Descriptor: | 4-[5-(1-methylpyrazol-4-yl)-3-[2-(1-methylpyrazol-4-yl)ethyl]imidazol-4-yl]benzenecarbonitrile, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reevaluation of bromodomain ligands targeting BAZ2A.
Protein Sci., 32, 2023
|
|
7BLB
 
 | | BAZ2A bromodomain in complex with GSK4027 chemical probe | | Descriptor: | 4-bromo-2-methyl-5-[[(3~{R},5~{R})-1-methyl-5-phenyl-piperidin-3-yl]amino]pyridazin-3-one, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reevaluation of bromodomain ligands targeting BAZ2A.
Protein Sci., 32, 2023
|
|
7BLD
 
 | | BAZ2A bromodomain in complex with compound UZH23 | | Descriptor: | 1-[3-(6-Methyl-2,3-dihydropyrazolo[5,1-b][1,3]oxazol-7-yl)indol-1-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Reevaluation of bromodomain ligands targeting BAZ2A.
Protein Sci., 32, 2023
|
|
6IEM
 
 | | Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, Argininosuccinate lyase, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
7BLA
 
 | | BAZ2A bromodomain in complex with TP-238 chemical probe | | Descriptor: | 6-{4-[3-(dimethylamino)propoxy]phenyl}-2-(methylsulfonyl)-N-[3-(1H-pyrazol-1-yl)propyl]pyrimidin-4-amine, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Reevaluation of bromodomain ligands targeting BAZ2A.
Protein Sci., 32, 2023
|
|
