6MQJ
 
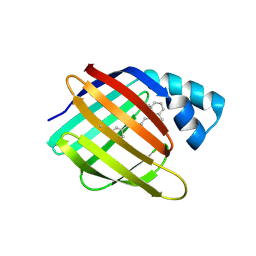 | |
5MPH
 
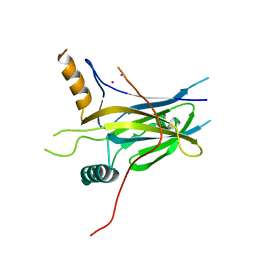 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | Grainyhead-like protein 1 homolog, IODIDE ION | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
3IKG
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
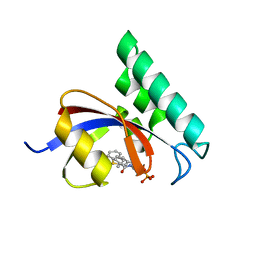
6MOI
 
 | | Dimeric DARPin C_angle_R5 complex with EpoR | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimeric DARPing CCR5 (C_angle_R5), ... | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C, Guo, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
5CBE
 
 | | E10 in complex with CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, C-X-C motif chemokine 13, E10 heavy chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
6B7G
 
 | | Solution NMR structure of BCoR in complex with AF9 (BCoR-AF9) | | Descriptor: | BCL-6 corepressor, Protein AF-9 | | Authors: | Schmidt, C.R, Kuntimaddi, A, Leach, B.I, Bushweller, J.H. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | BCOR Binding to MLL-AF9 Is Essential for Leukemia via Altered EYA1, SIX, and MYC Activity.
Blood Cancer Discov, 1, 2020
|
|
6B7R
 
 | | Truncated strand 11-less green fluorescent protein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Green fluorescent protein | | Authors: | Deng, A, Boxer, S.G. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Insight into the Photochemistry of Split Green Fluorescent Proteins: A Unique Role for a His-Tag.
J. Am. Chem. Soc., 140, 2018
|
|
6BDY
 
 | |
5MJ4
 
 | | INTERLEUKIN-23 COMPLEX WITH AN ANTAGONISTIC ALPHABODY, CRYSTAL FORM 2 | | Descriptor: | ALPHABODY MA12, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Desmet, J, Verstraete, K, Bloch, Y, Lorent, E, Wen, Y, Devreese, B, Vandenbroucke, K, Loverix, S, Hettmann, T, Deroo, S, Somers, K, Henderikx, P, Lasters, I, Savvides, S. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | STRUCTURAL BASIS OF IL-23 ANTAGONISM BY AN ALPHABODY PROTEIN
Nature Communications, 5, 2014
|
|
3IWU
 
 | | Crystal structure of Y116T/I16A double mutant of 5-hydroxyisourate hydrolase | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6MOQ
 
 | |
2BNU
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | T-CELL RECEPTOR ALPHA CHAIN C REGION, T-CELL RECEPTOR BETA CHAIN C REGION | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, Van Der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-04-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines.
J.Exp.Med., 201, 2005
|
|
5ML9
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CA5
 
 | | Structure of the C. elegans NONO-1 homodimer | | Descriptor: | 1,2-ETHANEDIOL, NONO-1 | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2015-06-29 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caenorhabditis elegans NONO-1: Insights into DBHS protein structure, architecture, and function.
Protein Sci., 24, 2015
|
|
5MP6
 
 | | Structure of the Unliganded Fab from HIV-1 Neutralizing Antibody CAP248-2B that Binds to the gp120 C-terminus - gp41 Interface, at two Angstrom resolution. | | Descriptor: | CAP248-2B Heavy Chain, CAP248-2B Light Chain, SULFATE ION | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog., 13, 2017
|
|
6MOP
 
 | |
6MPK
 
 | |
3IWZ
 
 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
6B7T
 
 | | Truncated strand 10-less green fluorescent protein | | Descriptor: | Green fluorescent protein,Green fluorescent protein | | Authors: | Deng, A, Boxer, S.G. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Insight into the Photochemistry of Split Green Fluorescent Proteins: A Unique Role for a His-Tag.
J. Am. Chem. Soc., 140, 2018
|
|
6B9J
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv138 | | Descriptor: | Fab vv138 Heavy chain, Fab vv138 Light chain, GLYCEROL, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
6MQY
 
 | |
7WCO
 
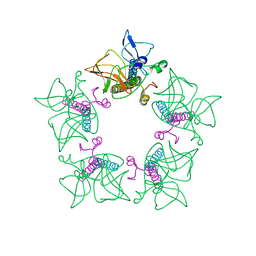 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
5MYC
 
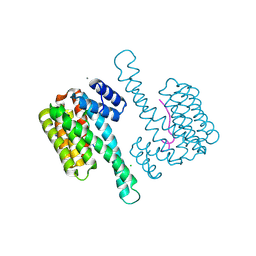 | | Crystal structure of human 14-3-3 sigma in complex with LRRK2 peptide pS910 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stevers, L.M, de Vries, R.M.J.M, Ottmann, C. | | Deposit date: | 2017-01-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural interface between LRRK2 and 14-3-3 protein.
Biochem. J., 474, 2017
|
|
