2HP5
 
 | | Crystal Structure of the OXA-10 W154G mutant at pH 7.0 | | Descriptor: | Beta-lactamase PSE-2, COBALT (II) ION, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP6
 
 | | Crystal structure of the OXA-10 W154A mutant at pH 7.5 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP9
 
 | | Crystal Structure of the OXA-10 W154A mutant at pH 6.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HPB
 
 | | Crystal structure of the OXA-10 W154A mutant at pH 9.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2GCC
 
 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
3GCC
 
 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1C74
 
 | | Structure of the double mutant (K53,56M) of phospholipase A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Tsai, M.D, Jain, M.K, Ramakumar, S. | | Deposit date: | 2000-01-22 | | Release date: | 2000-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the anionic interface preference and k*cat activation of pancreatic phospholipase A2.
Biochemistry, 39, 2000
|
|
3ET4
 
 | | Structure of Recombinant Haemophilus Influenzae E(P4) Acid Phosphatase | | Descriptor: | MAGNESIUM ION, Outer membrane protein P4, NADP phosphatase, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Recombinant Haemophilus Influenzae E (P4) Acid Phosphatase
Reveals a New Member of the Haloacid Dehalogenase Superfamily.
Biochemistry, 46, 2007
|
|
3ET5
 
 | |
9BHW
 
 | |
9BHT
 
 | | Septin Hexameric Complex SEPT2/SEPT6/SEPT7 of Ciona intestinalis by Cryo-EM | | Descriptor: | CiSeptin-2, CiSeptin-6, CiSeptin-7, ... | | Authors: | Mendonca, D.C, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2024-04-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Insights into Ciona intestinalis Septins: Complexes Suggest a Mechanism for Nucleotide-dependent Interfacial Cross-talk.
J.Mol.Biol., 436, 2024
|
|
4FRW
 
 | |
1BNO
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BNP
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
6A77
 
 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the Fab fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, Roundabout homolog 1 | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A76
 
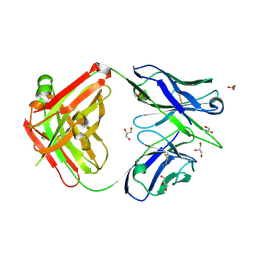 | | Crystal structure of the Fab fragment of B5209B, a murine monoclonal antibody specific for the fifth immunoglobulin domain (Ig5) of human ROBO1 | | Descriptor: | GLYCEROL, Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A79
 
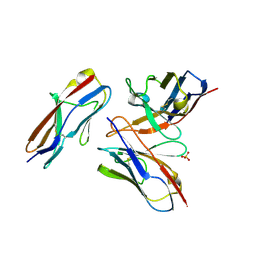 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the mutant scFv fragment (P103A) of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Yokota, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
8GQE
 
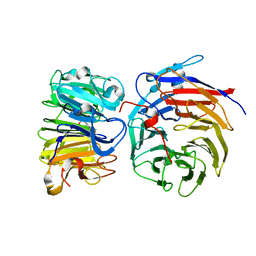 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
6A78
 
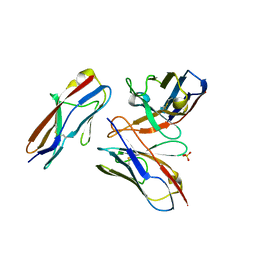 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the scFv fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain and linker region of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
1E3O
 
 | | Crystal structure of Oct-1 POU dimer bound to MORE | | Descriptor: | 5'-D(*AP*TP*GP*CP*AP*TP*GP*AP*GP*GP*A)-3', 5'-D(*TP*CP*CP*TP*CP*AP*TP*GP*CP*AP*T)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Schoeler, H, Wilmanns, M. | | Deposit date: | 2000-06-20 | | Release date: | 2001-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
1FEZ
 
 | | THE CRYSTAL STRUCTURE OF BACILLUS CEREUS PHOSPHONOACETALDEHYDE HYDROLASE COMPLEXED WITH TUNGSTATE, A PRODUCT ANALOG | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE HYDROLASE, TUNGSTATE(VI)ION | | Authors: | Morais, M.C, Zhang, W, Baker, A.S, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2000-07-24 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily.
Biochemistry, 39, 2000
|
|
7KC9
 
 | | Ricin bound to VHH antibody V5G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KD2
 
 | | Ricin bound to VHH antibody V11B2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KDU
 
 | | Ricin bound to VHH antibody V5E4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KBK
 
 | | Ricin bound to VHH antibody V6E11 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ricin, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
