1AK2
 
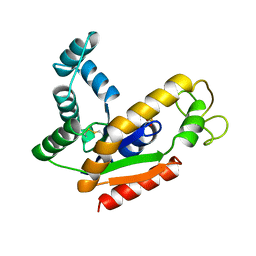 | | ADENYLATE KINASE ISOENZYME-2 | | Descriptor: | ADENYLATE KINASE ISOENZYME-2, SULFATE ION | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of bovine mitochondrial adenylate kinase: comparison with isoenzymes in other compartments.
Protein Sci., 5, 1996
|
|
1AK4
 
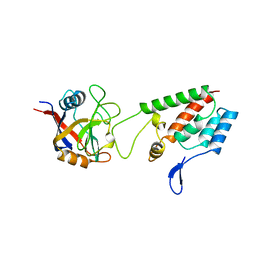 | | HUMAN CYCLOPHILIN A BOUND TO THE AMINO-TERMINAL DOMAIN OF HIV-1 CAPSID | | Descriptor: | CYCLOPHILIN A, HIV-1 CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Vajdos, F.F, Worthylake, D.K, Sundquist, W.I. | | Deposit date: | 1997-05-28 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid.
Cell(Cambridge,Mass.), 87, 1996
|
|
1AK5
 
 | |
1AK6
 
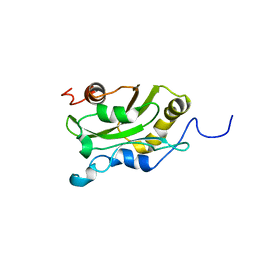 | | DESTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1AK7
 
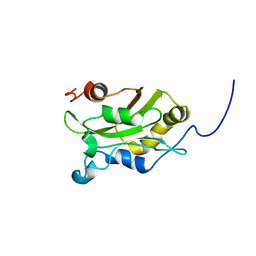 | | DESTRIN, NMR, 20 STRUCTURES | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1AK8
 
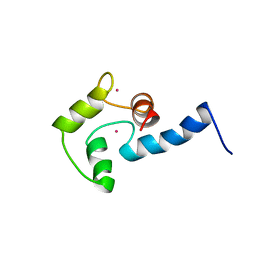 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | Descriptor: | CALMODULIN, CERIUM (III) ION | | Authors: | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
1AK9
 
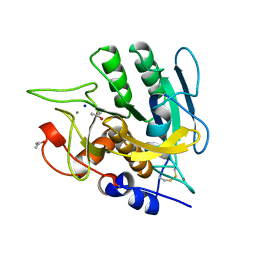 | | SUBTILISIN MUTANT 8321 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-05-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AKA
 
 | |
1AKB
 
 | |
1AKC
 
 | |
1AKD
 
 | | CYTOCHROME P450CAM FROM PSEUDOMONAS PUTIDA, COMPLEXED WITH 1S-CAMPHOR | | Descriptor: | CAMPHOR, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Schlichting, I, Jung, C, Schulze, H. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome P-450cam complexed with the (1S)-camphor enantiomer.
FEBS Lett., 415, 1997
|
|
1AKE
 
 | |
1AKG
 
 | | ALPHA-CONOTOXIN PNIB FROM CONUS PENNACEUS | | Descriptor: | ALPHA-CONOTOXIN PNIB | | Authors: | Hu, S.-H, Martin, J.L. | | Deposit date: | 1997-05-18 | | Release date: | 1998-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure at 1.1 A resolution of alpha-conotoxin PnIB: comparison with alpha-conotoxins PnIA and GI.
Biochemistry, 36, 1997
|
|
1AKH
 
 | | MAT A1/ALPHA2/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MATING-TYPE PROTEIN A-1), ... | | Authors: | Li, T, Jin, Y, Vershon, A.K, Wolberger, C. | | Deposit date: | 1997-05-19 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract.
Nucleic Acids Res., 26, 1998
|
|
1AKI
 
 | | THE STRUCTURE OF THE ORTHORHOMBIC FORM OF HEN EGG-WHITE LYSOZYME AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | LYSOZYME | | Authors: | Carter, D, He, J, Ruble, J.R, Wright, B. | | Deposit date: | 1997-05-19 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1AKJ
 
 | | COMPLEX OF THE HUMAN MHC CLASS I GLYCOPROTEIN HLA-A2 AND THE T CELL CORECEPTOR CD8 | | Descriptor: | BETA 2-MICROGLOBULIN, HIV REVERSE TRANSCRIPTASE EPITOPE, MHC CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), ... | | Authors: | Tormo, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-05-21 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2.
Nature, 387, 1997
|
|
1AKK
 
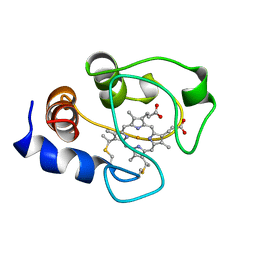 | | SOLUTION STRUCTURE OF OXIDIZED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Gray, H.B, Luchinat, C, Reddig, T, Rosato, A, Turano, P. | | Deposit date: | 1997-05-22 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized horse heart cytochrome c.
Biochemistry, 36, 1997
|
|
1AKL
 
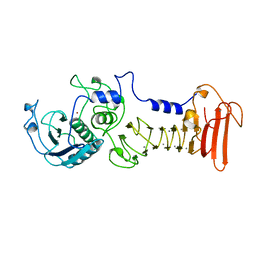 | | ALKALINE PROTEASE FROM PSEUDOMONAS AERUGINOSA IFO3080 | | Descriptor: | ALKALINE PROTEASE, CALCIUM ION, ZINC ION | | Authors: | Miyatake, H, Hata, Y, Fujii, T, Hamada, K, Morihara, K, Katsube, Y. | | Deposit date: | 1995-09-16 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the unliganded alkaline protease from Pseudomonas aeruginosa IFO3080 and its conformational changes on ligand binding.
J.Biochem.(Tokyo), 118, 1995
|
|
1AKM
 
 | |
1AKN
 
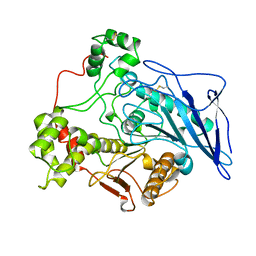 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-05-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
1AKO
 
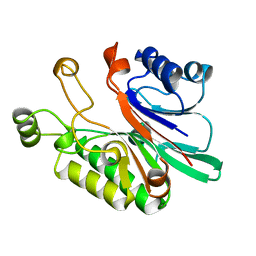 | | EXONUCLEASE III FROM ESCHERICHIA COLI | | Descriptor: | EXONUCLEASE III | | Authors: | Mol, C.D, Kuo, C.-F, Thayer, M.M, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the multifunctional DNA-repair enzyme exonuclease III.
Nature, 374, 1995
|
|
1AKP
 
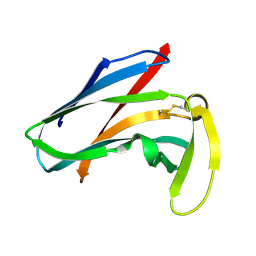 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
1AKQ
 
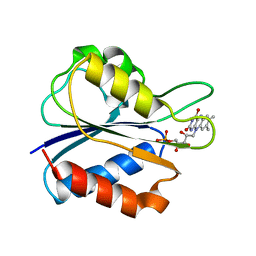 | | D95A OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-03-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKR
 
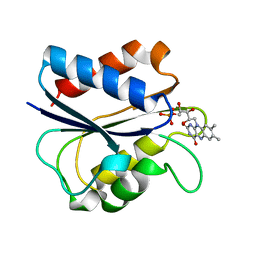 | | G61A OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AKS
 
 | | CRYSTAL STRUCTURE OF THE FIRST ACTIVE AUTOLYSATE FORM OF THE PORCINE ALPHA TRYPSIN | | Descriptor: | ALPHA TRYPSIN, CALCIUM ION | | Authors: | Johnson, A, Krishnaswamy, S, Sundaram, P.V, Pattabhi, V. | | Deposit date: | 1996-07-24 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure at 1.8 A resolution of an active autolysate form of porcine alpha-trysoin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
