7XLF
 
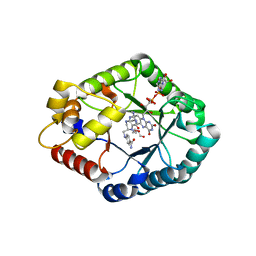 | | Crystal structure of CH3-THF complex of methylenetetrahydrofolate reductase from Sphingobium sp. SYK-6 | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yu, H.Y, Senda, M, Senda, T. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structural of methylenetetrahydrofolate reductase from Sphingobium sp. SYK-6
To Be Published
|
|
7XLD
 
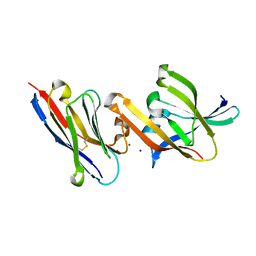 | | Crystal structure of IsdH linker-NEAT3 bound to a nanobody (VHH) | | Descriptor: | Iron-regulated surface determinant protein H, MAGNESIUM ION, Nanobody VHH6, ... | | Authors: | Caaveiro, J.M.M, Valenciano-Bellido, S, Tsumoto, K. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting hemoglobin receptors IsdH and IsdB of Staphylococcus aureus with a single VHH antibody inhibits bacterial growth.
J.Biol.Chem., 299, 2023
|
|
7XLB
 
 | |
7XL7
 
 | |
7XL6
 
 | |
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
7XL1
 
 | | Crystal structure of chimeric 7D12-Vob nanobody at 1.65 Angstrom | | Descriptor: | Chimeric 7D12-Vob nanobody, MALONATE ION | | Authors: | Caaveiro, J.M.M, Kinoshita, S, Mori, C, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
7XL0
 
 | | Crystal structure of Vobarilizumab at 1.70 Angstrom | | Descriptor: | GLYCEROL, Nanobody Vobarilizumab, SULFATE ION | | Authors: | Caaveiro, J.M.M, Mori, C, Kinoshita, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
7XKS
 
 | | Crystal structure of an alkaline pectate lyase from Bacillus clausii | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase | | Authors: | Zhou, C, Zheng, Y.Y, Liu, W.D, Ma, Y. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an Alkaline Pectate Lyase and Rational Engineering with Improved Thermo-Alkaline Stability for Efficient Ramie Degumming.
Int J Mol Sci, 24, 2022
|
|
7XKR
 
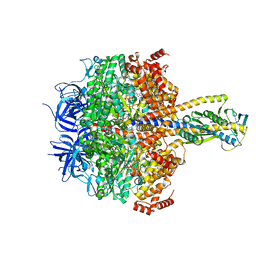 | | F1 domain of FoF1-ATPase with the up form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKQ
 
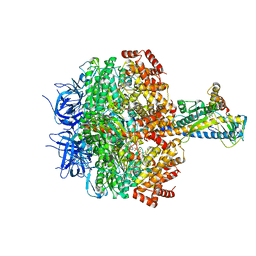 | | F1 domain of FoF1-ATPase with the down form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKP
 
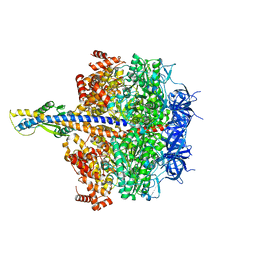 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKO
 
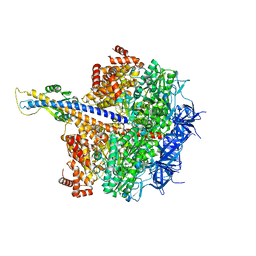 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,nucleotide depeleted | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKM
 
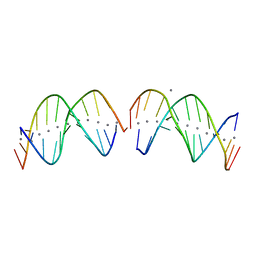 | |
7XKH
 
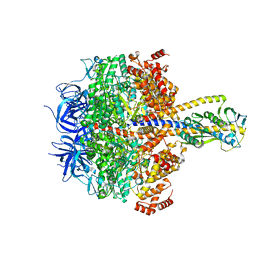 | | Nucleotide-depleted F1 domain of FoF1-ATPase from Bacillus PS3, state1 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XK9
 
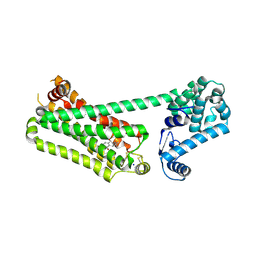 | | Structure of human beta2 adrenergic receptor bound to constrained isoproterenol | | Descriptor: | (5R,6R)-6-(propan-2-ylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
7XK7
 
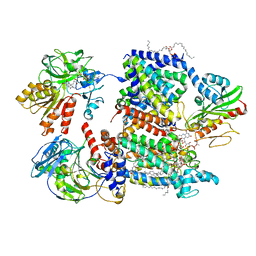 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, with korormicin | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK6
 
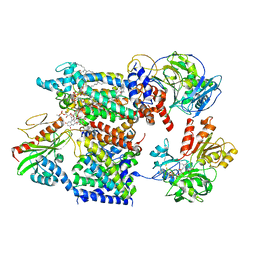 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, with aurachin D-42 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aurachin D, CALCIUM ION, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK5
 
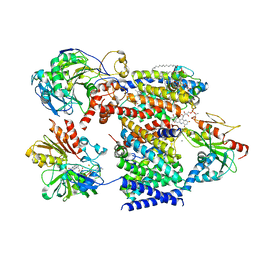 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 3 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK4
 
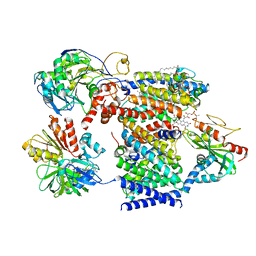 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK3
 
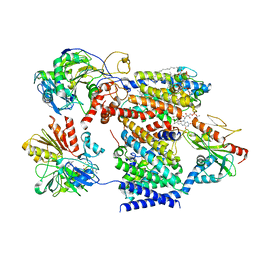 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 1 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7XK1
 
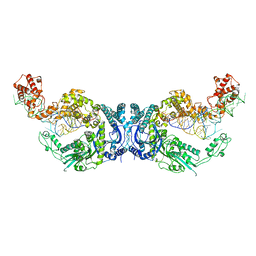 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK0
 
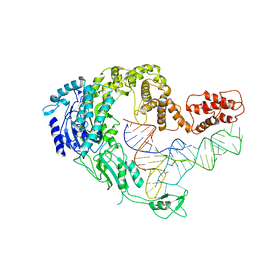 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XJZ
 
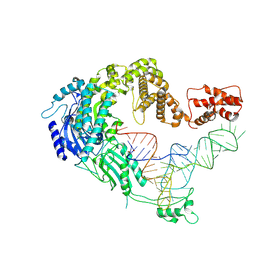 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA binding state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
