7AHQ
 
 | | Cryo-EM structure of F-actin stabilized by trans-optoJASP-8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Raunser, S. | | Deposit date: | 2020-09-25 | | Release date: | 2021-01-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Resolves Molecular Recognition Of An Optojasp Photoswitch Bound To Actin Filaments In Both Switch States.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3PUU
 
 | |
1DYH
 
 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | 5-DEAZAFOLIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
7AQ9
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583W | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
1DRH
 
 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sawaya, M.R, Kraut, J. | | Deposit date: | 1994-11-30 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
1DYI
 
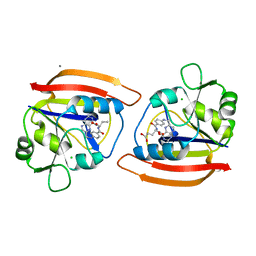 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
7AS8
 
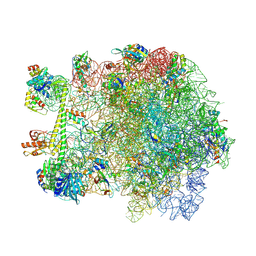 | | Bacillus subtilis ribosome quality control complex state B. Ribosomal 50S subunit with P-tRNA, RqcH, and RqcP/YabO | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for Bacterial Ribosome-Associated Quality Control by RqcH and RqcP.
Mol.Cell, 81, 2021
|
|
3PIO
 
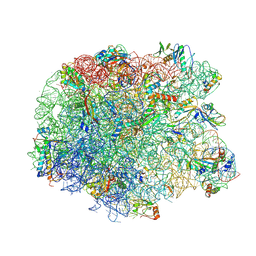 | | Crystal structure of the synergistic antibiotic pair lankamycin and lankacidin in complex with the large ribosomal subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Shapira, T, Bashan, A, Zimmerman, E, Arakawa, K, Kinashi, H, Rozenberg, H, Yonath, A. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2473 Å) | | Cite: | Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7AQ2
 
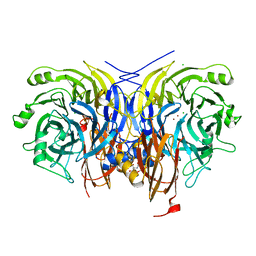 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
3PNG
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((2-fluoro-2-(3-fluorophenyl)ethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-2-fluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
3PUI
 
 | | Cocaine Esterase with mutations G4C, S10C | | Descriptor: | CHLORIDE ION, Cocaine esterase, SODIUM ION | | Authors: | Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2010-12-04 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Subunit stabilization and polyethylene glycolation of cocaine esterase improves in vivo residence time.
Mol.Pharmacol., 80, 2011
|
|
7B0Y
 
 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
8KH5
 
 | | Cryo-EM structure of the GPR174-Gs complex bound to endogenous lysoPS | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nie, Y, Qiu, Z, Zheng, S, Chen, S. | | Deposit date: | 2023-08-21 | | Release date: | 2023-10-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
7AUZ
 
 | | LTA4 hydrolase in complex with compound LYS006 | | Descriptor: | (3~{S})-3-azanyl-4-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
8KGK
 
 | | Cryo-EM structure of the GPR61-Gs complex | | Descriptor: | G-protein coupled receptor 61, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nie, Y, Qiu, Z, Zheng, S. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
7AV1
 
 | | LTA4 hydrolase in complex with fragment2 | | Descriptor: | 2-[5-(4-methoxyphenyl)-1,2,3,4-tetrazol-2-yl]ethanamine, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
1GUY
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | CADMIUM ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
7AV0
 
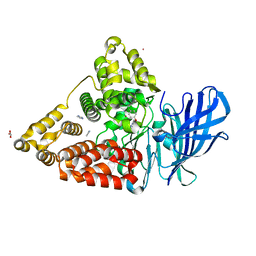 | | LTA4 hydrolase in complex with compound R(13) | | Descriptor: | (3~{R})-3-azanyl-4-[5-[4-(4-chloranylphenoxy)phenyl]-1,2,3,4-tetrazol-2-yl]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
8KH4
 
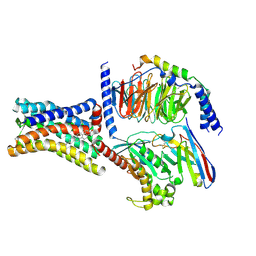 | | Cryo-EM structure of the GPR161-Gs complex | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nie, Y, Qiu, Z, Zheng, S, Chen, S. | | Deposit date: | 2023-08-21 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
7AV2
 
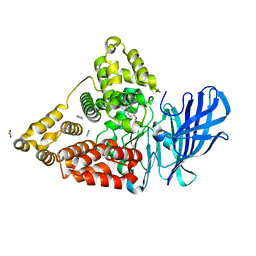 | | LTA4 hydrolase in complex with fragment1 | | Descriptor: | (4-phenoxyphenyl)methanol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
1GUZ
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
8OTZ
 
 | | 48-nm repeat of the native axonemal doublet microtubule from bovine sperm | | Descriptor: | ATP6V1F neighbor, CFAP97 domain containing 1, Chromosome 13 C20orf85 homolog, ... | | Authors: | Leung, M.R, Zeng, J, Zhang, R, Zeev-Ben-Mordehai, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural specializations of the sperm tail.
Cell, 186, 2023
|
|
3QIJ
 
 | | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R | | Descriptor: | Protein 4.1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Zhong, N, Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R
to be published
|
|
6ZNO
 
 | | The pointed end complex of dynactin with the p150 projection docked | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
