1VR2
 
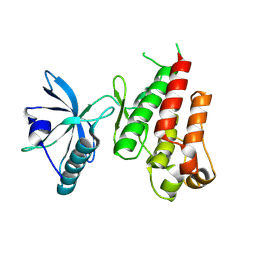 | | HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 (KDR) KINASE DOMAIN | | Descriptor: | PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR KINASE) | | Authors: | Mctigue, M, Wickersham, J, Pinko, C, Showalter, R, Parast, C, Tempczyk-Russell, A, Gehring, M, Mroczkowski, B, Kan, C, Villafranca, J, Appelt, K. | | Deposit date: | 1998-12-03 | | Release date: | 2000-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis.
Structure Fold.Des., 7, 1999
|
|
6C4I
 
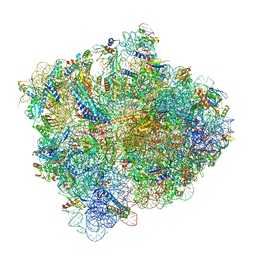 | |
2C6O
 
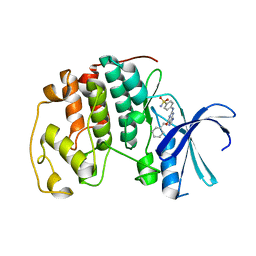 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
1S63
 
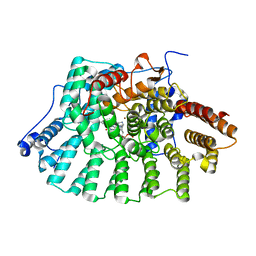 | | Human protein farnesyltransferase complexed with L-778,123 and FPP | | Descriptor: | 4-[(5-{[4-(3-CHLOROPHENYL)-3-OXOPIPERAZIN-1-YL]METHYL}-1H-IMIDAZOL-1-YL)METHYL]BENZONITRILE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis Reveals that Anticancer Clinical Candidate L-778,123 Inhibits Protein Farnesyltransferase and Geranylgeranyltransferase-I by Different Binding Modes.
Biochemistry, 43, 2004
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
2CJQ
 
 | |
7SNZ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((2-((4-cyanophenyl)amino)pyrimidin-4-yl)amino)-5,7-dimethylindolizine-2-carbonitrile (JLJ604) | | Descriptor: | (4R)-6-{[2-(4-cyanoanilino)pyrimidin-4-yl]amino}-5,7-dimethylindolizine-2-carbonitrile, 1,4-DIAMINOBUTANE, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
5MGP
 
 | | Structural basis for ArfA-RF2 mediated translation termination on stop-codon lacking mRNAs | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Mueller, C, Beckert, B, Arenz, S, Berninghausen, O, Beckmann, R, Wilson, N.D. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for ArfA-RF2-mediated translation termination on mRNAs lacking stop codons.
Nature, 541, 2017
|
|
7SO6
 
 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
1RWQ
 
 | | Human Dipeptidyl peptidase IV in complex with 5-aminomethyl-6-(2,4-dichloro-phenyl)-2-(3,5-dimethoxy-phenyl)-pyrimidin-4-ylamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(AMINOMETHYL)-6-(2,4-DICHLOROPHENYL)-2-(3,5-DIMETHOXYPHENYL)PYRIMIDIN-4-AMINE, Dipeptidyl peptidase IV | | Authors: | Hennig, M, Thoma, R, Stihle, M. | | Deposit date: | 2003-12-17 | | Release date: | 2004-12-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminomethylpyrimidines as novel DPP-IV inhibitors: a 10(5)-fold activity increase by optimization of aromatic substituents
Bioorg.Med.Chem.Lett., 14, 2004
|
|
7NQH
 
 | | 55S mammalian mitochondrial ribosome with mtRF1a and P-site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7SNP
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-(2-morpholinoethoxy)phenoxy)phenyl)acrylonitrile (JLJ530) | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-[2-(morpholin-4-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO2
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
5FWP
 
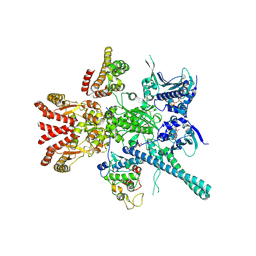 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Atomic Structure of Hsp90:Cdc37:Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
7SO1
 
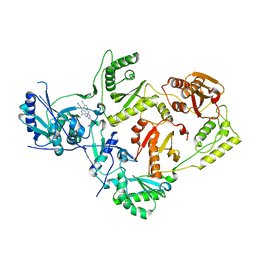 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.727 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
2C6T
 
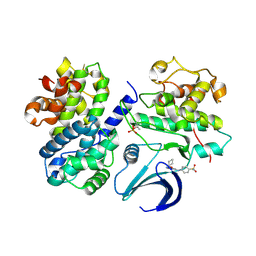 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-{[5-(CYCLOHEXYLOXY)[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL]AMINO}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-11 | | Release date: | 2005-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7SO3
 
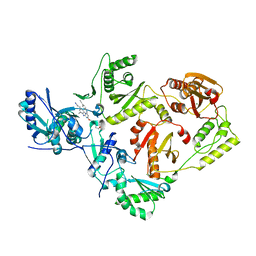 | | Crystal Structure of HIV-1 Reverse Transcriptase K103N/Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.767 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO4
 
 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7OF3
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
5FJV
 
 | | Crystal structure of the extracellular domain of alpha2 nicotinic acetylcholine receptor in pentameric assembly | | Descriptor: | EPIBATIDINE, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-2 | | Authors: | Giastas, P, Kouvatsos, N, Chroni-Tzartou, D, Tzartos, S.J. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a Human Neuronal Nachr Extracellular Domain in Pentameric Assembly: Ligand-Bound Alpah2 Homopentamer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LZB
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
7NSJ
 
 | | 55S mammalian mitochondrial ribosome with tRNA(P/P) and tRNA(E*) | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
6CQU
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase with Reactivator HI-6 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
5FKK
 
 | | TetR(D) N82A mutant in complex with anhydrotetracycline and magnesium | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Schneider, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modular Organisation of Inducer Recognition and Allostery in the Tetracycline Repressor
FEBS J., 283, 2016
|
|
5DUX
 
 | | Crystal structure of the human galectin-4 N-terminal carbohydrate recognition domain in complex with 2'-fucosyllactose | | Descriptor: | FORMIC ACID, GLYCEROL, Galectin-4, ... | | Authors: | Bum-Erdene, K, Blanchard, H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3'-sulfo-lactose, and 2'-fucosyllactose.
Sci Rep, 6, 2016
|
|
