2ZHI
 
 | |
3IG7
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFP with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-{1-[cis-3-(acetylamino)cyclobutyl]-1H-imidazol-4-yl}-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2Z7F
 
 | |
2V5W
 
 | | Crystal structure of HDAC8-substrate complex | | Descriptor: | GLYCYL-GLYCYL-GLYCINE, HISTONE DEACETYLASE 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
2ZR6
 
 | |
1OCT
 
 | | CRYSTAL STRUCTURE OF THE OCT-1 POU DOMAIN BOUND TO AN OCTAMER SITE: DNA RECOGNITION WITH TETHERED DNA-BINDING MODULES | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), PROTEIN (OCT-1 POU DOMAIN) | | Authors: | Klemm, J.D, Rould, M.A, Aurora, R, Herr, W, Pabo, C.O. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules.
Cell(Cambridge,Mass.), 77, 1994
|
|
2ZR4
 
 | |
2ZR5
 
 | |
2ZQS
 
 | |
3IGG
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFQ with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-[1-(cis-3-hydroxycyclobutyl)-1H-imidazol-4-yl]-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1OKV
 
 | | Cyclin A binding groove inhibitor H-Arg-Arg-Leu-Ile-Phe-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, H-ARG-ARG-LEU-ILE-PHE-NH2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-07-30 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1NMV
 
 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NOW
 
 | | Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
1OGU
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2/CYCLIN A COMPLEXED WITH A 2-ARYLAMINO-4-CYCLOHEXYLMETHYL-5-NITROSO-6-AMINOPYRIMIDINE INHIBITOR | | Descriptor: | 4-{[4-AMINO-6-(CYCLOHEXYLMETHOXY)-5-NITROSOPYRIMIDIN-2-YL]AMINO}BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-05-13 | | Release date: | 2003-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of 2-Arylamino-4-Cyclohexyl Methyl-5-Nitroso-6-Aminopyrimidine Inhibitors of Cyclin-Dependent Kinases 1 and 2
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1OIR
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 1-(DIMETHYLAMINO)-3-(4-{{4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)PYRIMIDIN-2-YL]AMINO}PHENOXY)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3TE5
 
 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3S2P
 
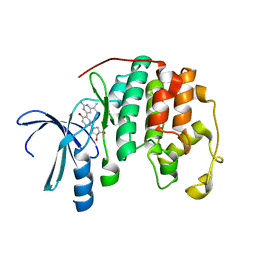 | | Crystal structure of CDK2 with a 2-aminopyrimidine compound | | Descriptor: | (3S,4S)-1-{3-[2-amino-6-(propan-2-yl)pyrimidin-4-yl]-4-hydroxyphenyl}pyrrolidine-3,4-diol, Cyclin-dependent kinase 2 | | Authors: | Kim, K.-H, Lee, J, Jeong, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of 2-aminopyrimidines as CDK1 and CDK2 inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RTR
 
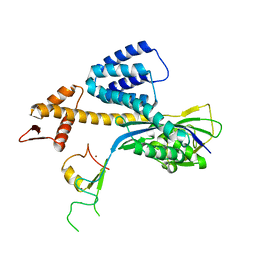 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RZB
 
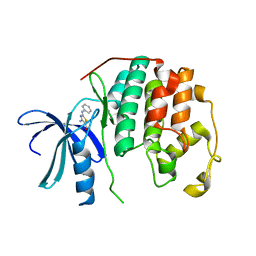 | | CDK2 in complex with inhibitor RC-2-23 | | Descriptor: | 4-amino-2-(phenylamino)-1,3-thiazole-5-carboxamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-05-11 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3S00
 
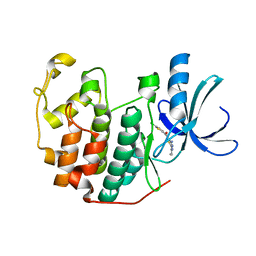 | | CDK2 in complex with inhibitor L4-14 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](5-chlorothiophen-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-05-12 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | Authors: | Skerlova, J, Pachl, P, Rezacova, P. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
1HCK
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HUMAN CYCLIN-DEPENDENT KINASE 2, MAGNESIUM ION | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
3SAY
 
 | | Crystal structure of human glycogen synthase kinase 3 beta (GSK3b) in complex with inhibitor 142 | | Descriptor: | (3Z)-N,N-diethyl-3-[(3E)-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-ylidene]-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide, (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, ... | | Authors: | Mazanetz, M.P, Cheng, R.K.Y, Rowan, F, Laughton, C.A, Barker, J.J, Fischer, P.M. | | Deposit date: | 2011-06-03 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Crystal structure of human glycogen synthase kinase 3 beta (GSK3b) in complex with inhibitor 142
To be Published
|
|
6ZVC
 
 | |
3SMK
 
 | | Crystal structure of human 14-3-3 sigma C38V/N166H in complex with TASK-3 peptide and stabilizer Cotylenin A | | Descriptor: | 14-3-3 protein sigma, Cotylenin A, MAGNESIUM ION, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem. Biol., 20, 2013
|
|
