3UWY
 
 | |
4AP4
 
 | | Rnf4 - ubch5a - ubiquitin heterotrimeric complex | | Descriptor: | E3 UBIQUITIN LIGASE RNF4, UBIQUITIN C, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Plechanovova, A, Hay, R.T, Tatham, M.H, Jaffray, E, Naismith, J.H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a Ring E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis
Nature, 489, 2012
|
|
1N0S
 
 | | ENGINEERED LIPOCALIN FLUA IN COMPLEX WITH FLUORESCEIN | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, Bilin-binding protein, SULFATE ION | | Authors: | Korndoerfer, I.P, Skerra, A. | | Deposit date: | 2002-10-15 | | Release date: | 2003-08-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of an "anticalin" with tailored specificity for fluorescein reveals high structural plasticity of the lipocalin loop region.
Proteins, 53, 2003
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3UWE
 
 | | AKR1C3 complexed with 3-phenoxybenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-phenoxybenzoic acid, Aldo-keto reductase family 1 member C3, ... | | Authors: | Jackson, V.J, Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-12-01 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of AKR1C3 with 3-phenoxybenzoic acid bound
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3UST
 
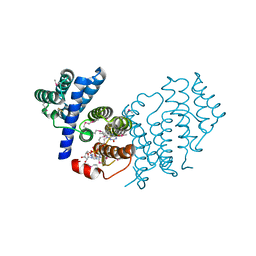 | | Structure of BmNPV ORF075 (p33) | | Descriptor: | AcMNPV orf92, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yuan, Y.A, Hou, Y, Xia, Q. | | Deposit date: | 2011-11-23 | | Release date: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Bombyx mori nucleopolyhedrovirus ORF75 reveals a pseudo-dimer of thiol oxidase domains with a putative substrate-binding pocket
J.Gen.Virol., 93, 2012
|
|
4AD5
 
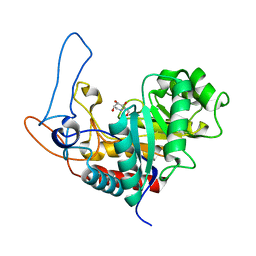 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-deoxymannojirimycin and alpha-1,2-mannobiose | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD1
 
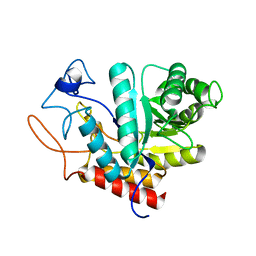 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens | | Descriptor: | GLYCOSYL HYDROLASE FAMILY 71 | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V1T
 
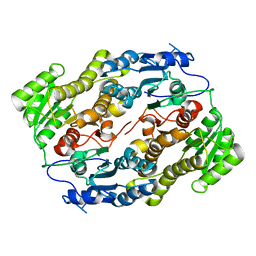 | |
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
1PQC
 
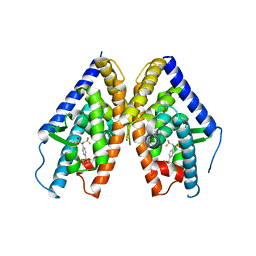 | | HUMAN LXR BETA HORMONE RECEPTOR COMPLEXED WITH T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Oxysterols receptor LXR-beta | | Authors: | Farnegardh, M, Bonn, T, Sun, S, Ljunggren, J, Ahola, H, Wilhelmsson, A, Gustafsson, J.-A, Carlquist, M. | | Deposit date: | 2003-06-18 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The three-dimensional structure of the liver X receptor beta reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands.
J.Biol.Chem., 278, 2003
|
|
4AKJ
 
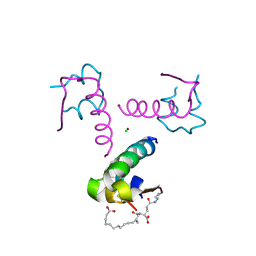 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4ALU
 
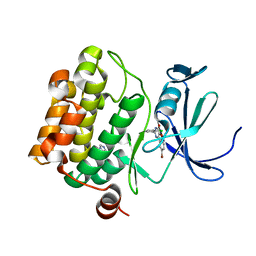 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-bromo-2-(2-chlorophenyl)[1]benzofuro[3,2-d]pyrimidin-4(3H)-one, IMIDAZOLE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7KPG
 
 | | Blocking Fab 25 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Heavy chain of Fab 25 anti-SIRP-alpha antibody, Light chain of Fab 25 anti-SIRP-alpha antibody, SULFATE ION, ... | | Authors: | Sim, J, Pons, J. | | Deposit date: | 2020-11-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Targeting the myeloid checkpoint receptor SIRP alpha potentiates innate and adaptive immune responses to promote anti-tumor activity.
J Hematol Oncol, 13, 2020
|
|
4A00
 
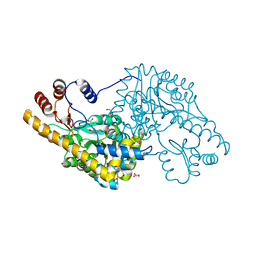 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
4A1T
 
 | | Co-Complex of the of NS3-4A protease with the inhibitory peptide CP5- 46-A (in-House data) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4ACL
 
 | | 3D Structure of DotU from Francisella novicida | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, SODIUM ION, ... | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2011-12-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Structure of the Conserved Type Six Secretion Protein Tssl (Dotu) from Francisella Novicida
J.Mol.Biol., 419, 2012
|
|
3VZ1
 
 | |
3ZFH
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | CHLORIDE ION, INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | Rao, V.A, Shepherd, S.M, Owen, R, Hunter, W.N. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Pseudomonas Aeruginosa Inosine 5'-Monophosphate Dehydrogenase
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3WYX
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 6-((3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-2-(cyclohexylamino)nicotinonitrile | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
4AI5
 
 | |
3VNW
 
 | | Crystal structure of cytochrome c552 from Thermus thermophilus at pH 5.44 | | Descriptor: | Cytochrome c-552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yamada, S, Bouley-Ford, N.D, Keller, G.E, Winkler, J.R, Gray, H.B. | | Deposit date: | 2012-01-18 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Snapshots of a protein folding intermediate
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VG3
 
 | | Cadmium derivative of human LFABP | | Descriptor: | CADMIUM ION, Fatty acid-binding protein, liver, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
4A3Q
 
 | | The 2.15 Angstrom resolution crystal structure of Staphylococcus aureus alanine racemase | | Descriptor: | ACETATE ION, ALANINE RACEMASE 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Scaletti, E.R, Luckner, S.R, Krause, K.L. | | Deposit date: | 2011-10-04 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Features and Kinetic Characterization of Alanine Racemase from Staphylococcus Aureus (Mu50)
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A0S
 
 | | STRUCTURE OF THE 2-OCTENOYL-COA CARBOXYLASE REDUCTASE CINF IN COMPLEX WITH NADP AND 2-OCTENOYL-COA | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A, OCTENOYL-COA REDUCTASE/CARBOXYLASE | | Authors: | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | Deposit date: | 2011-09-12 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual carbon fixation gives rise to diverse polyketide extender units.
Nat. Chem. Biol., 8, 2011
|
|
