7UHY
 
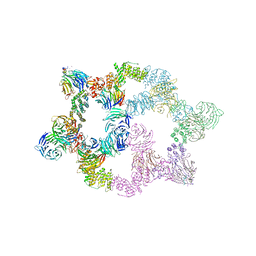 | | Human GATOR2 complex | | Descriptor: | GATOR complex protein MIOS, GATOR complex protein WDR24, GATOR complex protein WDR59, ... | | Authors: | Rogala, K.B, Valenstein, M.L, Lalgudi, P.V. | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of the nutrient-sensing hub GATOR2.
Nature, 607, 2022
|
|
4P1J
 
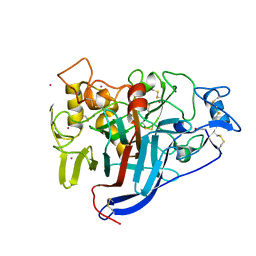 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, SAMARIUM (III) ION, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form
To Be Published
|
|
8D15
 
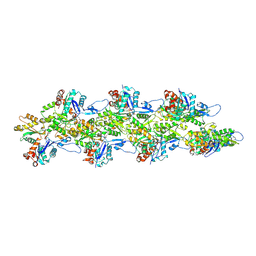 | | Bent ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
4YL8
 
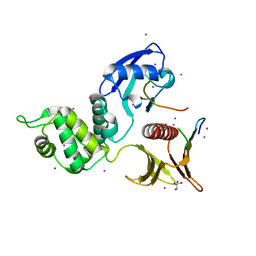 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
4YLQ
 
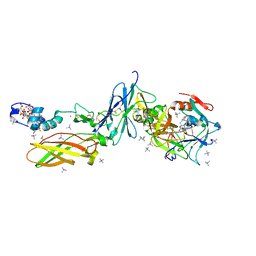 | | Crystal Structure of a FVIIa-Trypsin Chimera (FT) in Complex with Soluble Tissue Factor | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-PROPANOL, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity.
J.Biol.Chem., 291, 2016
|
|
4P1H
 
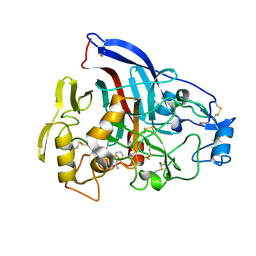 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, Exoglucanase 1, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form
to be published
|
|
8D17
 
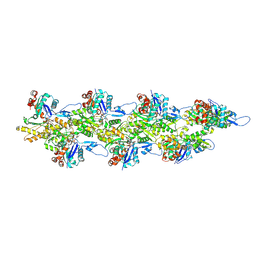 | | Straight ADP-F-actin 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
7UM6
 
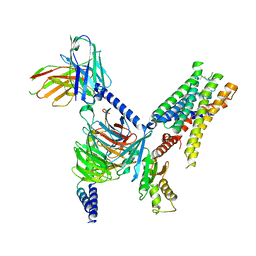 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | | Descriptor: | 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8D13
 
 | | Helical ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D16
 
 | | Bent ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D3Y
 
 | | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8F0E
 
 | |
2GBC
 
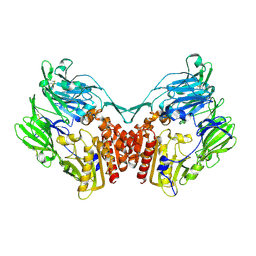 | | Native DPP-IV (CD26) from Rat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
8D3X
 
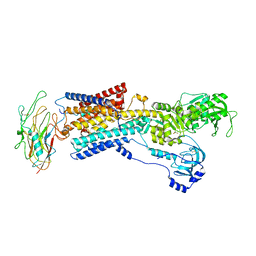 | | Human alpha3 Na+/K+-ATPase in its K+-occluded state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
7DVQ
 
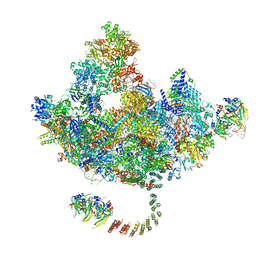 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
8D3U
 
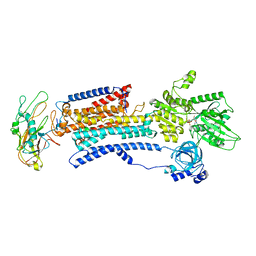 | | Human alpha3 Na+/K+-ATPase in its Na+-occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D4T
 
 | | Mammalian CIV with GDN bound | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Di Trani, J, Rubinstein, J. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian complex IV inhibition by steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8D3W
 
 | | Human alpha3 Na+/K+-ATPase in its AMPPCP-bound cytoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sodium/potassium-transporting ATPase subunit alpha-3, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
7E2Y
 
 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8D3V
 
 | | Human alpha3 Na+/K+-ATPase in its cytoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
7E2X
 
 | | Apo serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
