1NCJ
 
 | | N-CADHERIN, TWO-DOMAIN FRAGMENT | | Descriptor: | CALCIUM ION, PROTEIN (N-CADHERIN), URANYL (VI) ION | | Authors: | Tamura, K, Shan, W.-S, Hendrickson, W.A, Colman, D.R, Shapiro, L. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function analysis of cell adhesion by neural (N-) cadherin.
Neuron, 20, 1998
|
|
2CCD
 
 | |
4XPA
 
 | | X-ray structure of Drosophila dopamine transporter bound to 3,4dichlorophenethylamine | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
8HBD
 
 | | Cryo-EM structure of IRL1620-bound ETBR-Gi complex | | Descriptor: | Endothelin receptor type B,Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Ji, Y, Jiang, Y, Duan, J, Xu, H.E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
8HCX
 
 | | Cryo-EM structure of Endothelin1-bound ETBR-Gq complex | | Descriptor: | Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
2CE8
 
 | |
4XVA
 
 | | Crystal structure of wild type cytochrome c peroxidase | | Descriptor: | BENZIMIDAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
8HCQ
 
 | | Cryo-EM structure of endothelin1-bound ETAR-Gq complex | | Descriptor: | Endothelin-1, Endothelin-1 receptor,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
4XXB
 
 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
7UOH
 
 | | PRMT5/MEP50 crystal structure with MTA and an achiral, class 1, non-atropisomeric inhibitor bound | | Descriptor: | (2M)-2-[(4M)-4-{4-(aminomethyl)-1-oxo-8-[(2R)-oxolan-2-yl]-1,2-dihydrophthalazin-6-yl}-1-methyl-1H-pyrazol-5-yl]-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and evaluation of achiral, non-atropisomeric 4-(aminomethyl)phthalazin-1(2H)-one derivatives as novel PRMT5/MTA inhibitors.
Bioorg.Med.Chem., 71, 2022
|
|
5RZA
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z729352906 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-fluorophenyl)methyl]-1lambda~6~,2-thiazolidine-1,1-dione, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZN
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1745658474 | | Descriptor: | 1,2-ETHANEDIOL, 2-(trifluoromethyl)pyrimidine-5-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
4XQG
 
 | |
4XXK
 
 | | Crystal structure of the Semet-derivative of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | Descriptor: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | Authors: | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | Deposit date: | 2015-01-30 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5RYV
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1373430305 | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-[(oxan-4-yl)methyl]pyridin-2-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
4XYD
 
 | | Nitric oxide reductase from Roseobacter denitrificans (RdNOR) | | Descriptor: | CALCIUM ION, COPPER (II) ION, FE (III) ION, ... | | Authors: | Crow, A. | | Deposit date: | 2015-02-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Biochemistry, 55, 2016
|
|
5RYZ
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434868 | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-N-(2-fluorophenyl)piperidin-4-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZD
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z198194394 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-fluorophenyl)piperazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZS
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1703168683 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-fluoro-4-methylphenyl)methanesulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
6CEQ
 
 | |
8H4I
 
 | | DHA-bound FFAR4 in complex with Gs | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPU
 
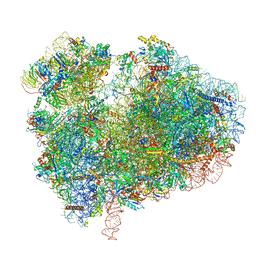 | | Cryo-EM structure of 80S ribosome from mouse kidney | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
