2X0L
 
 | | Crystal structure of a neuro-specific splicing variant of human histone lysine demethylase LSD1. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3 PEPTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Zibetti, C, Adamo, A, Binda, C, Forneris, F, Verpelli, C, Ginelli, E, Mattevi, A, Sala, C, Battaglioli, E. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative Splicing of the Histone Demethylase Lsd1/Kdm1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System.
J.Neurosci., 30, 2010
|
|
3BXF
 
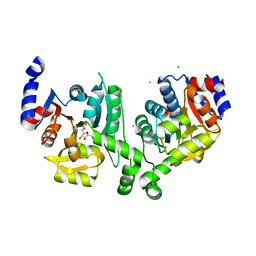 | | Crystal structure of effector binding domain of central glycolytic gene regulator (CggR) from Bacillus subtilis in complex with effector fructose-1,6-bisphosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, ... | | Authors: | Rezacova, P, Otwinowski, Z. | | Deposit date: | 2008-01-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol.Microbiol., 69, 2008
|
|
3BXG
 
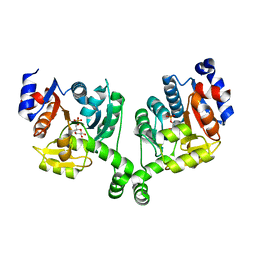 | |
3AQS
 
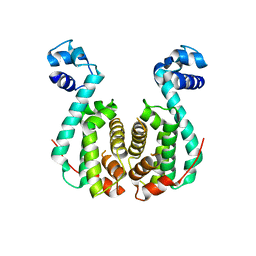 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3BXE
 
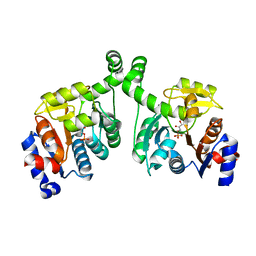 | |
4HPM
 
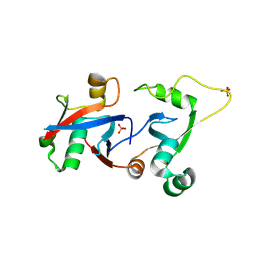 | | PCGF1 Ub fold (RAWUL)/BCORL1 PUFD Complex | | Descriptor: | BCL-6 corepressor-like protein 1, PHOSPHATE ION, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
3BXH
 
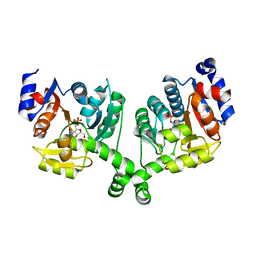 | |
1OKR
 
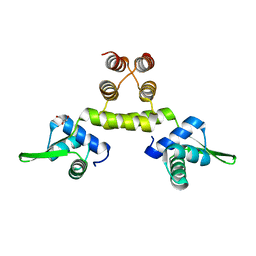 | | Three-dimensional structure of S.aureus methicillin-resistance regulating transcriptional repressor MecI. | | Descriptor: | CHLORIDE ION, GLYCEROL, METHICILLIN RESISTANCE REGULATORY PROTEIN MECI | | Authors: | Garcia-Castellanos, R, Marrero, A, Mallorqui-Fernandez, G, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2003-07-28 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Meci: Molecular Basis for Transcriptional Regulation of Staphylococcal Methicillin Resistance
J.Biol.Chem., 278, 2003
|
|
3OQ5
 
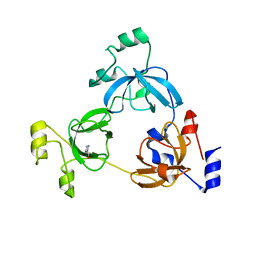 | | Crystal structure of the 3-MBT domain from human L3MBTL1 in complex with p53K382me1 | | Descriptor: | Cellular tumor antigen p53, Lethal(3)malignant brain tumor-like protein | | Authors: | Roy, S, West, L.E, Weiner, K.L, Hayashi, R, Shi, X, Appella, E, Gozani, O, Kutateladze, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5005 Å) | | Cite: | The MBT Repeats of L3MBTL1 Link SET8-mediated p53 Methylation at Lysine 382 to Target Gene Repression.
J.Biol.Chem., 285, 2010
|
|
4KUM
 
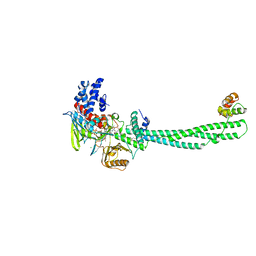 | | Structure of LSD1-CoREST-Tetrahydrofolate complex | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Calcutt, M.W, Newcomer, M.E, Wagner, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate.
Protein Sci., 23, 2014
|
|
3GLX
 
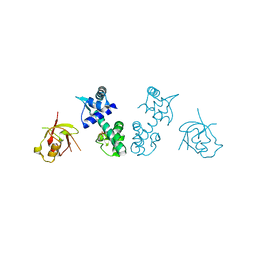 | | Crystal Structure Analysis of the DtxR(E175K) complexed with Ni(II) | | Descriptor: | Diphtheria toxin repressor, NICKEL (II) ION, PHOSPHATE ION | | Authors: | D'Aquino, J.A, Denninger, A, Moulin, A, D'Aquino, K.E, Ringe, D. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Decreased sensitivity to changes in the concentration of metal ions as the basis for the hyperactivity of DtxR(E175K).
J.Mol.Biol., 390, 2009
|
|
6XXS
 
 | |
6XYX
 
 | |
1DGD
 
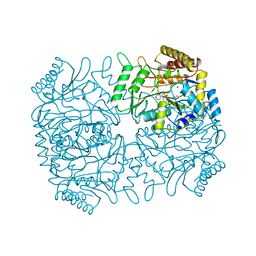 | |
1DGE
 
 | |
4C5G
 
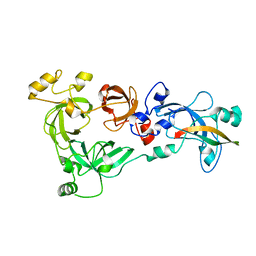 | |
4C5H
 
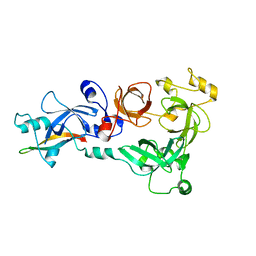 | |
4CIH
 
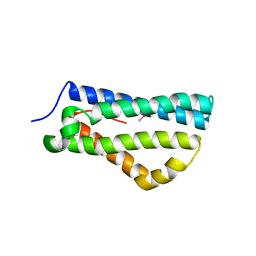 | | Structure of LntA-K180D-K181D from Listeria monocytogenes | | Descriptor: | LISTERIA NUCLEAR TARGETED PROTEIN A | | Authors: | Lebreton, A, Job, V, Ragon, M, Le Monnier, A, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2013-12-09 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA.
MBio, 5, 2014
|
|
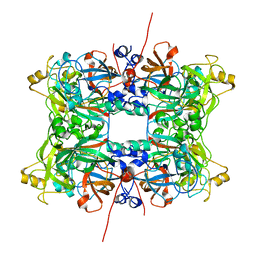
4C5E
 
 | |
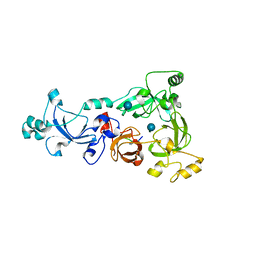
3H6Z
 
 | |
6TE1
 
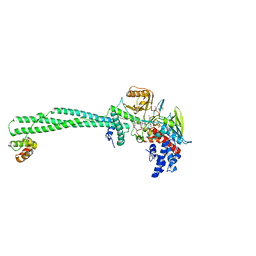 | | Structure of the KDM1A/CoREST complex with the inhibitor 2-[3-{4-chloro-3-[(4-chlorophenyl)ethynyl]phenyl}-1-(3-morpholin-4-ylpropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]-2-oxoethanol | | Descriptor: | 5-[4-cyclobutyl-1-[2-(4-piperidin-4-yloxyphenoxy)ethyl]imidazol-2-yl]-4-methyl-thieno[3,2-b]pyrrole, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Pasqualato, S, Cecatiello, V. | | Deposit date: | 2019-11-11 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of Reversible Inhibitors of KDM1A Efficacious in Acute Myeloid Leukemia Models.
Acs Med.Chem.Lett., 11, 2020
|
|
1AHD
 
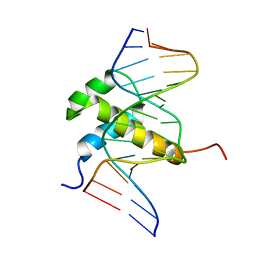 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
1JHF
 
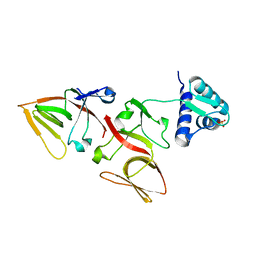 | | LEXA G85D MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JHH
 
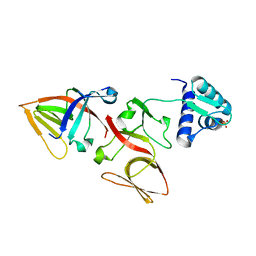 | | LEXA S119A MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
6ZVQ
 
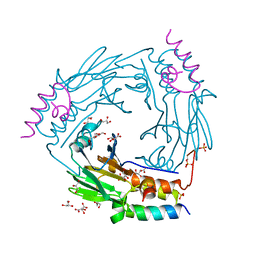 | | Complex between SMAD2 MH2 domain and peptide from Ski corepressor | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Mothers against decapentaplegic homolog 2, ... | | Authors: | Purkiss, A.G, Kjaer, S, George, R, Hill, C.S. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mutations in SKI in Shprintzen-Goldberg syndrome lead to attenuated TGF-beta responses through SKI stabilization.
Elife, 10, 2021
|
|
